HMGB1: roles in base excision repair and related function
- PMID: 20123074
- PMCID: PMC2818529
- DOI: 10.1016/j.bbagrm.2009.11.008
HMGB1: roles in base excision repair and related function
Abstract
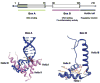
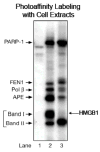
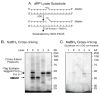
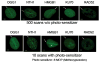
High mobility group box 1 (HMGB1) is a nonhistone architectural protein that is involved in many biological processes including chromatin remodeling, transcription, cell signaling of inflammation, DNA damage repair and others. Recent studies have identified the cross-link of HMGB1 with a DNA base excision repair intermediate indicating that this protein is involved in base excision repair (BER) pathway. Further characterization of the roles of HMGB1 in BER demonstrates that the protein acts as a cofactor to regulate BER sub-pathways by inhibiting single-nucleotide BER and stimulating long-patch BER through modulating the activities of base excision repair enzymes. Directing of base lesion repair to the long-patch sub-pathway can result in trinucleotide repeat instability suggesting an important role of HMGB1 in modulating genome stability.
Published by Elsevier B.V.
Figures






Similar articles
-
Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice.PLoS Genet. 2009 Dec;5(12):e1000749. doi: 10.1371/journal.pgen.1000749. Epub 2009 Dec 4. PLoS Genet. 2009. PMID: 19997493 Free PMC article.
-
Trinucleotide repeat instability via DNA base excision repair.DNA Repair (Amst). 2020 Sep;93:102912. doi: 10.1016/j.dnarep.2020.102912. DNA Repair (Amst). 2020. PMID: 33087278 Free PMC article. Review.
-
HMGB1 is a cofactor in mammalian base excision repair.Mol Cell. 2007 Sep 7;27(5):829-41. doi: 10.1016/j.molcel.2007.06.029. Mol Cell. 2007. PMID: 17803946 Free PMC article.
-
Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses.DNA Repair (Amst). 2019 Nov;83:102701. doi: 10.1016/j.dnarep.2019.102701. Epub 2019 Sep 16. DNA Repair (Amst). 2019. PMID: 31563843 Free PMC article. Review.
-
HMGB1 Stimulates Activity of Polymerase β on Nucleosome Substrates.Biochemistry. 2017 Jan 31;56(4):647-656. doi: 10.1021/acs.biochem.6b00569. Epub 2017 Jan 18. Biochemistry. 2017. PMID: 28098985 Free PMC article.
Cited by
-
Association of EGFR mutations and HMGB1 genetic polymorphisms in lung adenocarcinoma patients.J Cancer. 2019 Jun 2;10(13):2907-2914. doi: 10.7150/jca.31125. eCollection 2019. J Cancer. 2019. PMID: 31281467 Free PMC article.
-
High Mobility Group Box 1 and Cardiovascular Diseases: Study of Act and Connect.Cardiovasc Toxicol. 2024 Nov;24(11):1268-1286. doi: 10.1007/s12012-024-09919-5. Epub 2024 Sep 6. Cardiovasc Toxicol. 2024. PMID: 39242448 Review.
-
Emerging roles of the nucleolus in regulating the DNA damage response: the noncanonical DNA repair enzyme APE1/Ref-1 as a paradigmatical example.Antioxid Redox Signal. 2014 Feb 1;20(4):621-39. doi: 10.1089/ars.2013.5491. Epub 2013 Sep 21. Antioxid Redox Signal. 2014. PMID: 23879289 Free PMC article. Review.
-
HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction.Chromosoma. 2012 Aug;121(4):419-31. doi: 10.1007/s00412-012-0373-x. Epub 2012 Apr 28. Chromosoma. 2012. PMID: 22544226
-
The role of HMGB1/RAGE/TLR4 signaling pathways in cigarette smoke-induced inflammation in chronic obstructive pulmonary disease.Immun Inflamm Dis. 2022 Nov;10(11):e711. doi: 10.1002/iid3.711. Immun Inflamm Dis. 2022. PMID: 36301039 Free PMC article. Review.
References
-
- Reeck GR, Isackson PJ, Teller DC. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982;300:76–78. - PubMed
-
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. - PubMed
-
- Shirakawa H, Tsuda K, Yoshida M. Primary structure of non-histone chromosomal protein HMG2 revealed by the nucleotide sequence. Biochemistry. 1990;29:4419–4423. - PubMed
-
- Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

