Mammalian nuclear transplantation to Germinal Vesicle stage Xenopus oocytes - a method for quantitative transcriptional reprogramming
- PMID: 20123126
- PMCID: PMC2877800
- DOI: 10.1016/j.ymeth.2010.01.035
Mammalian nuclear transplantation to Germinal Vesicle stage Xenopus oocytes - a method for quantitative transcriptional reprogramming
Abstract
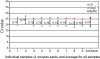
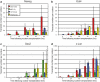
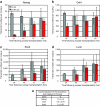
Full-grown Xenopus oocytes in first meiotic prophase contain an immensely enlarged nucleus, the Germinal Vesicle (GV), that can be injected with several hundred somatic cell nuclei. When the nuclei of mammalian somatic cells or cultured cell lines are injected into a GV, a wide range of genes that are not transcribed in the donor cells, including pluripotency genes, start to be transcriptionally activated, and synthesize primary transcripts continuously for several days. Because of the large size and abundance of Xenopus laevis oocytes, this experimental system offers an opportunity to understand the mechanisms by which somatic cell nuclei can be reprogrammed to transcribe genes characteristic of oocytes and early embryos. The use of mammalian nuclei ensures that there is no background of endogenous maternal transcripts of the kind that are induced. The induced gene transcription takes place in the absence of cell division or DNA synthesis and does not require protein synthesis. Here we summarize new as well as established results that characterize this experimental system. In particular, we describe optimal conditions for transplanting somatic nuclei to oocytes and for the efficient activation of transcription by transplanted nuclei. We make a quantitative determination of transcript numbers for pluripotency and housekeeping genes, comparing cultured somatic cell nuclei with those of embryonic stem cells. Surprisingly we find that the transcriptional activation of somatic nuclei differs substantially from one donor cell-type to another and in respect of different pluripotency genes. We also determine the efficiency of an injected mRNA translation into protein.
Figures






Similar articles
-
Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation.Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5483-8. doi: 10.1073/pnas.1000599107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212135 Free PMC article.
-
Widespread transcription in an amphibian oocyte relates to its reprogramming activity on transplanted somatic nuclei.Stem Cells Dev. 2012 Jan 20;21(2):181-90. doi: 10.1089/scd.2011.0162. Epub 2011 Jun 15. Stem Cells Dev. 2012. PMID: 21504359 Free PMC article.
-
Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes.Curr Biol. 2003 Jul 15;13(14):1206-13. doi: 10.1016/s0960-9822(03)00462-7. Curr Biol. 2003. PMID: 12867031
-
Using oocyte nuclei for studies on chromatin structure and gene expression.Methods. 2010 May;51(1):157-64. doi: 10.1016/j.ymeth.2010.02.002. Epub 2010 Feb 6. Methods. 2010. PMID: 20138999 Review.
-
Injected amphibian oocytes: a living test tube for the study of eukaryotic gene transcription?Biochem Soc Symp. 1977;(42):181-91. Biochem Soc Symp. 1977. PMID: 339919 Review.
Cited by
-
Epigenetic rejuvenation.Genes Cells. 2012 May;17(5):337-43. doi: 10.1111/j.1365-2443.2012.01595.x. Epub 2012 Apr 4. Genes Cells. 2012. PMID: 22487104 Free PMC article. Review.
-
Efficiencies and mechanisms of nuclear reprogramming.Cold Spring Harb Symp Quant Biol. 2010;75:189-200. doi: 10.1101/sqb.2010.75.002. Epub 2010 Nov 3. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21047900 Free PMC article.
-
Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process?Nat Rev Mol Cell Biol. 2011 Jun 23;12(7):453-9. doi: 10.1038/nrm3140. Nat Rev Mol Cell Biol. 2011. PMID: 21697902 Free PMC article. Review.
-
Histone variant macroH2A confers resistance to nuclear reprogramming.EMBO J. 2011 May 6;30(12):2373-87. doi: 10.1038/emboj.2011.144. EMBO J. 2011. PMID: 21552206 Free PMC article.
-
Initiation and maintenance of pluripotency gene expression in the absence of cohesin.Genes Dev. 2015 Jan 1;29(1):23-38. doi: 10.1101/gad.251835.114. Genes Dev. 2015. PMID: 25561493 Free PMC article.
References
-
- Callan H.G. Proc. Royal Soc. Lond. B: Biol. Sci. 1982;214:417–448. - PubMed
-
- Davidson E.H. Academic Press; New York: 1986. Gene Activity in Early Development.
-
- Woodland H.R., Flynn J.M., Wyllie A.J. Cell. 1979;18:165–171. - PubMed
-
- Elsdale T.R., Fischberg M., Smith S. Exp. Cell Res. 1958;14:642–643. - PubMed
-
- Gurdon J.B. Nature. 1974;248:772–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

