DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model
- PMID: 20123730
- PMCID: PMC2853144
- DOI: 10.1093/nar/gkq021
DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model
Abstract
RNA pseudoknots are functional structure elements with key roles in viral and cellular processes. Prediction of a pseudoknotted minimum free energy structure is an NP-complete problem. Practical algorithms for RNA structure prediction including restricted classes of pseudoknots suffer from high runtime and poor accuracy for longer sequences. A heuristic approach is to search for promising pseudoknot candidates in a sequence and verify those. Afterwards, the detected pseudoknots can be further analysed using bioinformatics or laboratory techniques. We present a novel pseudoknot detection method called DotKnot that extracts stem regions from the secondary structure probability dot plot and assembles pseudoknot candidates in a constructive fashion. We evaluate pseudoknot free energies using novel parameters, which have recently become available. We show that the conventional probability dot plot makes a wide class of pseudoknots including those with bulged stems manageable in an explicit fashion. The energy parameters now become the limiting factor in pseudoknot prediction. DotKnot is an efficient method for long sequences, which finds pseudoknots with higher accuracy compared to other known prediction algorithms. DotKnot is accessible as a web server at http://dotknot.csse.uwa.edu.au.
Figures
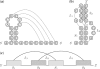
 and
and  and three loops
and three loops  ,
,  and
and  . (c) A pseudoknot has at least two crossing stems, which can be displayed as intervals on the line.
. (c) A pseudoknot has at least two crossing stems, which can be displayed as intervals on the line.
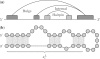
 interrupted by a bulge loop and an internal loop. (b) The corresponding secondary structure is shown.
interrupted by a bulge loop and an internal loop. (b) The corresponding secondary structure is shown.
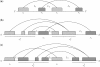
 and
and  are crossing. (b) Stem
are crossing. (b) Stem  and stem
and stem  are crossing. (c) Stem
are crossing. (c) Stem  and stem
and stem  are crossing.
are crossing.Similar articles
-
Heuristic RNA pseudoknot prediction including intramolecular kissing hairpins.RNA. 2011 Jan;17(1):27-38. doi: 10.1261/rna.2394511. Epub 2010 Nov 22. RNA. 2011. PMID: 21098139 Free PMC article.
-
Predicting pseudoknotted structures across two RNA sequences.Bioinformatics. 2012 Dec 1;28(23):3058-65. doi: 10.1093/bioinformatics/bts575. Epub 2012 Oct 8. Bioinformatics. 2012. PMID: 23044552 Free PMC article.
-
KnotSeeker: heuristic pseudoknot detection in long RNA sequences.RNA. 2008 Apr;14(4):630-40. doi: 10.1261/rna.968808. Epub 2008 Feb 26. RNA. 2008. PMID: 18314500 Free PMC article.
-
Pseudoknots in RNA Structure Prediction.Curr Protoc. 2023 Feb;3(2):e661. doi: 10.1002/cpz1.661. Curr Protoc. 2023. PMID: 36779804 Review.
-
Structure and function of pseudoknots involved in gene expression control.Wiley Interdiscip Rev RNA. 2014 Nov-Dec;5(6):803-22. doi: 10.1002/wrna.1247. Epub 2014 Jul 8. Wiley Interdiscip Rev RNA. 2014. PMID: 25044223 Free PMC article. Review.
Cited by
-
Molecular characterization of totiviruses in Xanthophyllomyces dendrorhous.Virol J. 2012 Jul 28;9:140. doi: 10.1186/1743-422X-9-140. Virol J. 2012. PMID: 22838956 Free PMC article.
-
Characterization of a Novel Megabirnavirus from Sclerotinia sclerotiorum Reveals Horizontal Gene Transfer from Single-Stranded RNA Virus to Double-Stranded RNA Virus.J Virol. 2015 Aug;89(16):8567-79. doi: 10.1128/JVI.00243-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063429 Free PMC article.
-
Gene from a novel plant virus satellite from grapevine identifies a viral satellite lineage.Virus Genes. 2013 Aug;47(1):114-8. doi: 10.1007/s11262-013-0921-3. Epub 2013 May 24. Virus Genes. 2013. PMID: 23703624
-
A Novel RNA Virus Related to Sobemoviruses Confers Hypovirulence on the Phytopathogenic Fungus Sclerotinia sclerotiorum.Viruses. 2019 Aug 16;11(8):759. doi: 10.3390/v11080759. Viruses. 2019. PMID: 31426425 Free PMC article.
-
A Novel Hypovirus Species From Xylariaceae Fungi Infecting Avocado.Front Microbiol. 2018 May 8;9:778. doi: 10.3389/fmicb.2018.00778. eCollection 2018. Front Microbiol. 2018. PMID: 29867781 Free PMC article.
References
-
- Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. - PubMed
-
- Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. - PubMed
-
- Tinoco I, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. - PubMed
-
- Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr. Opin. Struct. Biol. 2007;17:157–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

