Intronic elements in the Na+/I- symporter gene (NIS) interact with retinoic acid receptors and mediate initiation of transcription
- PMID: 20123735
- PMCID: PMC2879507
- DOI: 10.1093/nar/gkq023
Intronic elements in the Na+/I- symporter gene (NIS) interact with retinoic acid receptors and mediate initiation of transcription
Abstract
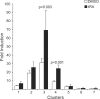
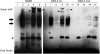
Activity of the sodium/iodide symporter (NIS) in lactating breast is essential for iodide (I(-)) accumulation in milk. Significant NIS upregulation was also reported in breast cancer, indicating a potential use of radioiodide treatment. All-trans-retinoic acid (tRA) is a potent ligand that enhances NIS expression in a subset of breast cancer cell lines and in experimental breast cancer models. Indirect tRA stimulation of NIS in breast cancer cells is very well documented; however, direct upregulation by tRA-activated nuclear receptors has not been identified yet. Aiming to uncover cis-acting elements directly regulating NIS expression, we screened evolutionary-conserved non-coding genomic sequences for responsiveness to tRA in MCF-7. Here, we report that a potent enhancer in the first intron of NIS mediates direct regulation by tRA-stimulated nuclear receptors. In vitro as well as in vivo DNA-protein interaction assays revealed direct association between retinoic acid receptor-alpha (RARalpha) and retinoid-X-receptor (RXR) with this enhancer. Moreover, using chromatin immunoprecipitation (ChIP) we uncovered early events of NIS transcription in response to tRA, which require the interaction of several novel intronic tRA responsive elements. These findings indicate a complex interplay between nuclear receptors, RNA Pol-II and multiple intronic RAREs in NIS gene, and they establish a novel mechanistic model for tRA-induced gene transcription.
Figures





Similar articles
-
Differential regulation of sodium/iodide symporter gene expression by nuclear receptor ligands in MCF-7 breast cancer cells.Endocrinology. 2005 Jul;146(7):3059-69. doi: 10.1210/en.2004-1334. Epub 2005 Apr 7. Endocrinology. 2005. PMID: 15817668
-
Activation of the PI3 kinase pathway by retinoic acid mediates sodium/iodide symporter induction and iodide transport in MCF-7 breast cancer cells.Cancer Res. 2009 Apr 15;69(8):3443-50. doi: 10.1158/0008-5472.CAN-08-3234. Epub 2009 Apr 7. Cancer Res. 2009. PMID: 19351850 Free PMC article.
-
Unliganded estrogen receptor-alpha activates transcription of the mammary gland Na+/I- symporter gene.Biochem Biophys Res Commun. 2006 Jul 14;345(4):1487-96. doi: 10.1016/j.bbrc.2006.05.049. Epub 2006 May 15. Biochem Biophys Res Commun. 2006. PMID: 16730657
-
Enhancement of sodium/iodide symporter expression in thyroid and breast cancer.Endocr Relat Cancer. 2006 Sep;13(3):797-826. doi: 10.1677/erc.1.01143. Endocr Relat Cancer. 2006. PMID: 16954431 Review.
-
Rexinoids and breast cancer prevention.Clin Cancer Res. 2007 Oct 15;13(20):5983-7. doi: 10.1158/1078-0432.CCR-07-1065. Clin Cancer Res. 2007. PMID: 17947457 Review. No abstract available.
Cited by
-
Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq.Genome Res. 2012 Dec;22(12):2385-98. doi: 10.1101/gr.135707.111. Epub 2012 Jul 26. Genome Res. 2012. PMID: 22835905 Free PMC article.
-
The sodium/iodide symporter NIS is a transcriptional target of the p53-family members in liver cancer cells.Cell Death Dis. 2013 Sep 19;4(9):e807. doi: 10.1038/cddis.2013.302. Cell Death Dis. 2013. PMID: 24052075 Free PMC article.
-
The Role of Iodine for Thyroid Function in Lactating Women and Infants.Endocr Rev. 2022 May 12;43(3):469-506. doi: 10.1210/endrev/bnab029. Endocr Rev. 2022. PMID: 35552681 Free PMC article. Review.
-
The Thyroid Na+/I- Symporter: Molecular Characterization and Genomic Regulation.Mol Imaging Radionucl Ther. 2017 Feb 9;26(Suppl 1):92-101. doi: 10.4274/2017.26.suppl.11. Mol Imaging Radionucl Ther. 2017. PMID: 28117294 Free PMC article.
-
The Transcription Factor Elf3 Is Essential for a Successful Mesenchymal to Epithelial Transition.Cells. 2019 Aug 9;8(8):858. doi: 10.3390/cells8080858. Cells. 2019. PMID: 31404945 Free PMC article.
References
-
- Spitzweg C, Joba W, Eisenmenger W, Heufelder AE. Analysis of human sodium iodide symporter gene expression in extrathyroidal tissues and cloning of its complementary deoxyribonucleic acids from salivary gland, mammary gland, and gastric mucosa. J. Clin. Endocrinol. Metab. 1998;83:1746–1751. - PubMed
-
- Tazebay UH, Wapnir IL, Levy O, Dohan O, Zuckier LS, Zhao QH, Deng HF, Amenta PS, Fineberg S, Pestell RG, et al. The mammary gland iodide transporter is expressed during lactation and in breast cancer. Nat. Med. 2000;6:871–878. - PubMed
-
- Stubbe P, Schulte FJ, Heidemann P. Iodine deficiency and brain development. Bibl. Nutr. Dieta. 1986:206–208. - PubMed
-
- Cho JY, Leveille R, Kao R, Rousset B, Parlow AF, Burak W.E., Jr, Mazzaferri EL, Jhiang SM. Hormonal regulation of radioiodide uptake activity and Na+/I- symporter expression in mammary glands. J. Clin. Endocrinol. Metab. 2000;85:2936–2943. - PubMed
-
- Perron B, Rodriguez AM, Leblanc G, Pourcher T. Cloning of the mouse sodium iodide symporter and its expression in the mammary gland and other tissues. J. Endocrinol. 2001;170:185–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

