Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding
- PMID: 20123966
- PMCID: PMC2838068
- DOI: 10.1128/MCB.01602-09
Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding
Abstract
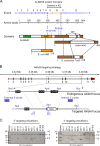
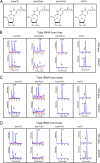
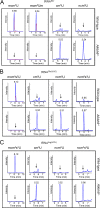
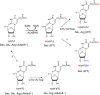
Uridines in the wobble position of tRNA are almost invariably modified. Modifications can increase the efficiency of codon reading, but they also prevent mistranslation by limiting wobbling. In mammals, several tRNAs have 5-methoxycarbonylmethyluridine (mcm5U) or derivatives thereof in the wobble position. Through analysis of tRNA from Alkbh8-/- mice, we show here that ALKBH8 is a tRNA methyltransferase required for the final step in the biogenesis of mcm5U. We also demonstrate that the interaction of ALKBH8 with a small accessory protein, TRM112, is required to form a functional tRNA methyltransferase. Furthermore, prior ALKBH8-mediated methylation is a prerequisite for the thiolation and 2'-O-ribose methylation that form 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) and 5-methoxycarbonylmethyl-2'-O-methyluridine (mcm5Um), respectively. Despite the complete loss of all of these uridine modifications, Alkbh8-/- mice appear normal. However, the selenocysteine-specific tRNA (tRNASec) is aberrantly modified in the Alkbh8-/- mice, and for the selenoprotein Gpx1, we indeed observed reduced recoding of the UGA stop codon to selenocysteine.
Figures




Similar articles
-
Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA.Nucleic Acids Res. 2011 Sep 1;39(17):7688-701. doi: 10.1093/nar/gkr406. Epub 2011 Jun 7. Nucleic Acids Res. 2011. PMID: 21653555 Free PMC article.
-
Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification.Am J Hum Genet. 2019 Jun 6;104(6):1202-1209. doi: 10.1016/j.ajhg.2019.03.026. Epub 2019 May 9. Am J Hum Genet. 2019. PMID: 31079898 Free PMC article.
-
Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage.PLoS One. 2015 Jul 6;10(7):e0131335. doi: 10.1371/journal.pone.0131335. eCollection 2015. PLoS One. 2015. PMID: 26147969 Free PMC article.
-
Genome recoding by tRNA modifications.Open Biol. 2016 Dec;6(12):160287. doi: 10.1098/rsob.160287. Open Biol. 2016. PMID: 27974624 Free PMC article. Review.
-
Elongator-a tRNA modifying complex that promotes efficient translational decoding.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):401-408. doi: 10.1016/j.bbagrm.2017.11.006. Epub 2017 Nov 21. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29170010 Review.
Cited by
-
Lysine methylation of VCP by a member of a novel human protein methyltransferase family.Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. Nat Commun. 2012. PMID: 22948820
-
Pathways to disease from natural variations in human cytoplasmic tRNAs.J Biol Chem. 2019 Apr 5;294(14):5294-5308. doi: 10.1074/jbc.REV118.002982. Epub 2019 Jan 14. J Biol Chem. 2019. PMID: 30643023 Free PMC article. Review.
-
Oligodendrocyte differentiation alters tRNA modifications and codon optimality-mediated mRNA decay.Nat Commun. 2022 Aug 25;13(1):5003. doi: 10.1038/s41467-022-32766-3. Nat Commun. 2022. PMID: 36008413 Free PMC article.
-
The epitranscriptomic writer ALKBH8 drives tolerance and protects mouse lungs from the environmental pollutant naphthalene.Epigenetics. 2020 Oct;15(10):1121-1138. doi: 10.1080/15592294.2020.1750213. Epub 2020 Apr 17. Epigenetics. 2020. PMID: 32303148 Free PMC article.
-
Methylated nucleosides in tRNA and tRNA methyltransferases.Front Genet. 2014 May 23;5:144. doi: 10.3389/fgene.2014.00144. eCollection 2014. Front Genet. 2014. PMID: 24904644 Free PMC article. Review.
References
-
- Carlson, B. A., M. E. Moustafa, A. Sengupta, U. Schweizer, R. Shrimali, M. Rao, N. Zhong, S. Wang, L. Feigenbaum, B. J. Lee, V. N. Gladyshev, and D. L. Hatfield. 2007. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 282:32591-32602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
