SRC induces podoplanin expression to promote cell migration
- PMID: 20123990
- PMCID: PMC2843215
- DOI: 10.1074/jbc.M109.047696
SRC induces podoplanin expression to promote cell migration
Abstract
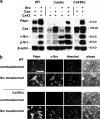
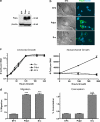
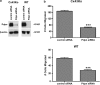
Nontransformed cells can force tumor cells to assume a normal morphology and phenotype by the process of contact normalization. Transformed cells must escape this process to become invasive and malignant. However, mechanisms underlying contact normalization have not been elucidated. Here, we have identified genes that are affected by contact normalization of Src-transformed cells. Tumor cells must migrate to become invasive and malignant. Src must phosphorylate the adaptor protein Cas (Crk-associated substrate) to promote tumor cell motility. We report here that Src utilizes Cas to induce podoplanin (Pdpn) expression to promote tumor cell migration. Pdpn is a membrane-bound extracellular glycoprotein that associates with endogenous ligands to promote tumor cell migration leading to cancer invasion and metastasis. In fact, Pdpn expression accounted for a major part of the increased migration seen in Src-transformed cells. Moreover, nontransformed cells suppressed Pdpn expression in adjacent Src-transformed cells. Of >39,000 genes, Pdpn was one of only 23 genes found to be induced by transforming Src activity and suppressed by contact normalization of Src-transformed cells. In addition, we found 16 genes suppressed by Src and induced by contact normalization. These genes encode growth factor receptors, adaptor proteins, and products that have not yet been annotated and may play important roles in tumor cell growth and migration.
Figures



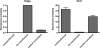
Similar articles
-
Heterocellular N-cadherin junctions enable nontransformed cells to inhibit the growth of adjacent transformed cells.Cell Commun Signal. 2022 Feb 17;20(1):19. doi: 10.1186/s12964-021-00817-9. Cell Commun Signal. 2022. PMID: 35177067 Free PMC article.
-
Independent effects of Src kinase and podoplanin on anchorage independent cell growth and migration.Mol Carcinog. 2022 Jul;61(7):677-689. doi: 10.1002/mc.23410. Epub 2022 Apr 26. Mol Carcinog. 2022. PMID: 35472679 Free PMC article.
-
Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration.Oncogene. 2009 Dec 3;28(48):4272-83. doi: 10.1038/onc.2009.278. Epub 2009 Sep 21. Oncogene. 2009. PMID: 19767772
-
Src and podoplanin forge a path to destruction.Drug Discov Today. 2019 Jan;24(1):241-249. doi: 10.1016/j.drudis.2018.07.009. Epub 2018 Aug 2. Drug Discov Today. 2019. PMID: 30077780 Review.
-
New concepts regarding focal adhesion kinase promotion of cell migration and proliferation.J Cell Biochem. 2006 Sep 1;99(1):35-52. doi: 10.1002/jcb.20956. J Cell Biochem. 2006. PMID: 16823799 Review.
Cited by
-
Antibody and lectin target podoplanin to inhibit oral squamous carcinoma cell migration and viability by distinct mechanisms.Oncotarget. 2015 Apr 20;6(11):9045-60. doi: 10.18632/oncotarget.3515. Oncotarget. 2015. PMID: 25826087 Free PMC article.
-
Plant lectin can target receptors containing sialic acid, exemplified by podoplanin, to inhibit transformed cell growth and migration.PLoS One. 2012;7(7):e41845. doi: 10.1371/journal.pone.0041845. Epub 2012 Jul 23. PLoS One. 2012. PMID: 22844530 Free PMC article.
-
Podoplanin--a novel marker in oral carcinogenesis.Tumour Biol. 2014 Sep;35(9):8407-13. doi: 10.1007/s13277-014-2266-5. Epub 2014 Jun 27. Tumour Biol. 2014. PMID: 24964963 Review.
-
Articular chondrocyte network mediated by gap junctions: role in metabolic cartilage homeostasis.Ann Rheum Dis. 2015 Jan;74(1):275-84. doi: 10.1136/annrheumdis-2013-204244. Epub 2013 Nov 13. Ann Rheum Dis. 2015. PMID: 24225059 Free PMC article.
-
SRC points the way to biomarkers and chemotherapeutic targets.Genes Cancer. 2012 May;3(5-6):426-35. doi: 10.1177/1947601912458583. Genes Cancer. 2012. PMID: 23226580 Free PMC article.
References
-
- Rubin H. (2006) Bioessays 28, 515–524 - PubMed
-
- Rubin H. (2008) Adv. Cancer Res. 100, 159–202 - PubMed
-
- Yeatman T. J. (2004) Nat. Rev. Cancer 4, 470–480 - PubMed
-
- Honda H., Oda H., Nakamoto T., Honda Z., Sakai R., Suzuki T., Saito T., Nakamura K., Nakao K., Ishikawa T., Katsuki M., Yazaki Y., Hirai H. (1998) Nat. Genet. 19, 361–365 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

