Proliferation of aneuploid human cells is limited by a p53-dependent mechanism
- PMID: 20123995
- PMCID: PMC2819684
- DOI: 10.1083/jcb.200905057
Proliferation of aneuploid human cells is limited by a p53-dependent mechanism
Abstract
Most solid tumors are aneuploid, and it has been proposed that aneuploidy is the consequence of an elevated rate of chromosome missegregation in a process called chromosomal instability (CIN). However, the relationship of aneuploidy and CIN is unclear because the proliferation of cultured diploid cells is compromised by chromosome missegregation. The mechanism for this intolerance of nondiploid genomes is unknown. In this study, we show that in otherwise diploid human cells, chromosome missegregation causes a cell cycle delay with nuclear accumulation of the tumor suppressor p53 and the cyclin kinase inhibitor p21. Deletion of the p53 gene permits the accumulation of nondiploid cells such that CIN generates cells with aneuploid genomes that resemble many human tumors. Thus, the p53 pathway plays an important role in limiting the propagation of aneuploid human cells in culture to preserve the diploid karyotype of the population. These data fit with the concordance of aneuploidy and disruption of the p53 pathway in many tumors, but the presence of aneuploid cells in some normal human and mouse tissues indicates that there are known exceptions to the involvement of p53 in aneuploid cells and that tissue context may be important in how cells respond to aneuploidy.
Figures
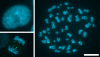

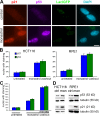

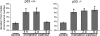

References
-
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. 2002. Molecular Biology of the Cell. Fourth edition Garland Science, New York: 1548 pp
-
- Baker S.J., Preisinger A.C., Jessup J.M., Paraskeva C., Markowitz S., Willson J.K., Hamilton S., Vogelstein B. 1990. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 50:7717–7722 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

