Mutual regulation of cyclin-dependent kinase and the mitotic exit network
- PMID: 20123997
- PMCID: PMC2819678
- DOI: 10.1083/jcb.200911128
Mutual regulation of cyclin-dependent kinase and the mitotic exit network
Abstract
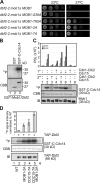
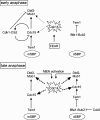
The mitotic exit network (MEN) is a spindle pole body (SPB)-associated, GTPase-driven signaling cascade that controls mitotic exit. The inhibitory Bfa1-Bub2 GTPase-activating protein (GAP) only associates with the daughter SPB (dSPB), raising the question as to how the MEN is regulated on the mother SPB (mSPB). Here, we show mutual regulation of cyclin-dependent kinase 1 (Cdk1) and the MEN. In early anaphase Cdk1 becomes recruited to the mSPB depending on the activity of the MEN kinase Cdc15. Conversely, Cdk1 negatively regulates binding of Cdc15 to the mSPB. In addition, Cdk1 phosphorylates the Mob1 protein to inhibit the activity of Dbf2-Mob1 kinase that regulates Cdc14 phosphatase. Our data revise the understanding of the spatial regulation of the MEN. Although MEN activity in the daughter cells is controlled by Bfa1-Bub2, Cdk1 inhibits MEN activity at the mSPB. Consistent with this model, only triple mutants that lack BUB2 and the Cdk1 phosphorylation sites in Mob1 and Cdc15 show mitotic exit defects.
Figures







Similar articles
-
Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit.J Cell Biol. 2006 Jan 30;172(3):335-46. doi: 10.1083/jcb.200507162. J Cell Biol. 2006. PMID: 16449187 Free PMC article.
-
The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit.Curr Biol. 2000 Nov 2;10(21):1379-82. doi: 10.1016/s0960-9822(00)00779-x. Curr Biol. 2000. PMID: 11084339
-
Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis.Curr Biol. 2001 Mar 6;11(5):345-50. doi: 10.1016/s0960-9822(01)00095-1. Curr Biol. 2001. PMID: 11267871
-
Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1.Small GTPases. 2015 Oct 2;6(4):196-201. doi: 10.1080/21541248.2015.1109023. Small GTPases. 2015. PMID: 26507466 Free PMC article. Review.
-
The Multiple Roles of the Cdc14 Phosphatase in Cell Cycle Control.Int J Mol Sci. 2020 Jan 21;21(3):709. doi: 10.3390/ijms21030709. Int J Mol Sci. 2020. PMID: 31973188 Free PMC article. Review.
Cited by
-
Zds1 regulates PP2A(Cdc55) activity and Cdc14 activation during mitotic exit through its Zds_C motif.J Cell Sci. 2012 Jun 15;125(Pt 12):2875-84. doi: 10.1242/jcs.097865. Epub 2012 Mar 16. J Cell Sci. 2012. PMID: 22427694 Free PMC article.
-
Meiotic Cytokinesis in Saccharomyces cerevisiae: Spores That Just Need Closure.J Fungi (Basel). 2024 Feb 6;10(2):132. doi: 10.3390/jof10020132. J Fungi (Basel). 2024. PMID: 38392804 Free PMC article. Review.
-
Monitoring spindle orientation: Spindle position checkpoint in charge.Cell Div. 2010 Dec 11;5:28. doi: 10.1186/1747-1028-5-28. Cell Div. 2010. PMID: 21143992 Free PMC article.
-
Proteome-wide forced interactions reveal a functional map of cell-cycle phospho-regulation in S. cerevisiae.Nucleus. 2024 Dec;15(1):2420129. doi: 10.1080/19491034.2024.2420129. Epub 2024 Dec 1. Nucleus. 2024. PMID: 39618027 Free PMC article.
-
Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis.Mol Cell Biol. 2011 Feb;31(4):721-35. doi: 10.1128/MCB.00403-10. Epub 2010 Dec 6. Mol Cell Biol. 2011. PMID: 21135117 Free PMC article.
References
-
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M.N. 2003. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 116:4119–4130 10.1242/jcs.00706 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

