Combination of V106I and V179D polymorphic mutations in human immunodeficiency virus type 1 reverse transcriptase confers resistance to efavirenz and nevirapine but not etravirine
- PMID: 20124001
- PMCID: PMC2849364
- DOI: 10.1128/AAC.01480-09
Combination of V106I and V179D polymorphic mutations in human immunodeficiency virus type 1 reverse transcriptase confers resistance to efavirenz and nevirapine but not etravirine
Abstract
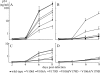
Etravirine (ETV) is a second-generation nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) introduced recently for salvage antiretroviral treatment after the emergence of NNRTI-resistant human immunodeficiency virus type 1 (HIV-1). Following its introduction, two naturally occurring mutations in HIV-1 RT, V106I and V179D, were listed as ETV resistance-associated mutations. However, the effect of these mutations on the development of NNRTI resistance has not been analyzed yet. To select highly NNRTI-resistant HIV-1 in vitro, monoclonal HIV-1 strains harboring V106I and V179D (HIV-1(V106I) and HIV-1(V179D)) were propagated in the presence of increasing concentrations of efavirenz (EFV). Interestingly, V179D emerged in one of three selection experiments from HIV-1(V106I) and V106I emerged in two of three experiments from HIV-1(V179D). Analysis of recombinant HIV-1 clones showed that the combination of V106I and V179D conferred significant resistance to EFV and nevirapine (NVP) but not to ETV. Structural analysis indicated that ETV can overcome the repulsive interactions caused by the combination of V106I and V179D through fine-tuning of its binding module to RT facilitated by its plastic structure, whereas EFV and NVP cannot because of their rigid structures. Analysis of clinical isolates showed comparable drug susceptibilities, and the same combination of mutations was found in some database patients who experienced virologic NNRTI-based treatment failure. The combination of V106I and V179D is a newly identified NNRTI resistance pattern of mutations. The combination of polymorphic and minor resistance-associated mutations should be interpreted carefully.
Figures


References
-
- Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski.2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. - PMC - PubMed
-
- Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers.2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. - PMC - PubMed
-
- Butler, I. F., I. Pandrea, P. A. Marx, and C. Apetrei.2007. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 5:23-45. - PubMed
-
- Das, K., A. D. Clark, Jr., P. J. Lewi, J. Heeres, M. R. De Jonge, L. M. Koymans, H. M. Vinkers, F. Daeyaert, D. W. Ludovici, M. J. Kukla, B. De Corte, R. W. Kavash, C. Y. Ho, H. He, M. A. Lichtenstein, K. Andries, R. Pauwels, M. P. De Bethune, P. L. Boyer, P. Clark, S. H. Hughes, P. A. Janssen, and E. Arnold.2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550-2560. - PubMed
-
- Duan, Y., C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. Kollman.2003. A point-charged force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999-2012. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

