DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA
- PMID: 20124025
- PMCID: PMC2865930
- DOI: 10.1534/genetics.110.113969
DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA
Abstract
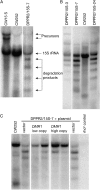
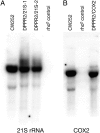
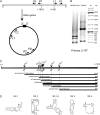
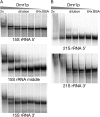
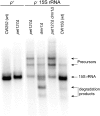
Pentatricopeptide repeat (PPR) proteins form the largest known RNA-binding protein family and are found in all eukaryotes, being particularly abundant in higher plants. PPR proteins localize mostly in mitochondria and chloroplasts, where they modulate organellar genome expression on the post-transcriptional level. The Saccharomyces cerevisiae DMR1 (CCM1, YGR150C) encodes a PPR protein that localizes to mitochondria. Deletion of DMR1 results in a complete and irreversible loss of respiratory capacity and loss of wild-type mtDNA by conversion to rho(-)/rho(0) petites, regardless of the presence of introns in mtDNA. The phenotype of the dmr1Delta mitochondria is characterized by fragmentation of the small subunit mitochondrial rRNA (15S rRNA), that can be reversed by wild-type Dmr1p. Other mitochondrial transcripts, including the large subunit mitochondrial rRNA (21S rRNA), are not affected by the lack of Dmr1p. The purified Dmr1 protein specifically binds to different regions of 15S rRNA in vitro, consistent with the deletion phenotype. Dmr1p is therefore the first yeast PPR protein, which has an rRNA target and is probably involved in the biogenesis of mitochondrial ribosomes and translation.
Figures

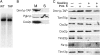
Similar articles
-
Yeast pentatricopeptide protein Dmr1 (Ccm1) binds a repetitive AU-rich motif in the small subunit mitochondrial ribosomal RNA.RNA. 2020 Sep;26(9):1268-1282. doi: 10.1261/rna.074880.120. Epub 2020 May 28. RNA. 2020. PMID: 32467310 Free PMC article.
-
Two independent activities define Ccm1p as a moonlighting protein in Saccharomyces cerevisiae.Biosci Rep. 2012 Dec;32(6):549-57. doi: 10.1042/BSR20120066. Biosci Rep. 2012. PMID: 22861139 Free PMC article.
-
Ccm1p is a 15S rRNA primary transcript processing factor as elucidated by a novel in vivo system in Saccharomyces cerevisiae.Curr Genet. 2020 Aug;66(4):775-789. doi: 10.1007/s00294-020-01064-0. Epub 2020 Mar 9. Curr Genet. 2020. PMID: 32152734 Free PMC article.
-
Human pentatricopeptide proteins: only a few and what do they do?RNA Biol. 2013;10(9):1433-8. doi: 10.4161/rna.24770. Epub 2013 Apr 23. RNA Biol. 2013. PMID: 23635806 Free PMC article. Review.
-
Analysis of mitochondrial biogenesis and protein localization by genetic screens and automated imaging.Methods Enzymol. 2024;706:97-123. doi: 10.1016/bs.mie.2024.07.022. Epub 2024 Aug 15. Methods Enzymol. 2024. PMID: 39455236 Review.
Cited by
-
When nuclear-encoded proteins and mitochondrial RNAs do not get along, species split apart.EMBO Rep. 2017 Jan;18(1):8-10. doi: 10.15252/embr.201643645. Epub 2016 Dec 16. EMBO Rep. 2017. PMID: 27986790 Free PMC article.
-
Mitochondrial ribosome assembly in health and disease.Cell Cycle. 2015;14(14):2226-50. doi: 10.1080/15384101.2015.1053672. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030272 Free PMC article. Review.
-
Global analysis of Saccharomyces cerevisiae growth in mucin.G3 (Bethesda). 2021 Oct 19;11(11):jkab294. doi: 10.1093/g3journal/jkab294. G3 (Bethesda). 2021. PMID: 34849793 Free PMC article.
-
Mitochondrial mRNA and the small subunit rRNA in budding yeasts undergo 3'-end processing at conserved species-specific elements.RNA. 2025 Jan 22;31(2):208-223. doi: 10.1261/rna.080254.124. RNA. 2025. PMID: 39572231 Free PMC article.
-
Yeast model analysis of novel polymerase gamma variants found in patients with autosomal recessive mitochondrial disease.Hum Genet. 2015 Sep;134(9):951-66. doi: 10.1007/s00439-015-1578-x. Epub 2015 Jun 16. Hum Genet. 2015. PMID: 26077851 Free PMC article.
References
-
- Andres, C., C. Lurin and I. D. Sluyter, 2007. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129 14–22.
-
- Blatch, G. L., and M. Lassle, 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21 932–939. - PubMed
-
- Bonnefoy, N., and T. D. Fox, 2002. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Enzymol. 350 97–111. - PubMed
-
- Bousquet, I., G. Dujardin, R. O. Poyton and P. P. Slonimski, 1990. Two group I mitochondrial introns in the cob-box and coxI genes require the same MRS1/PET157 nuclear gene product for splicing. Curr. Genet. 18 117–124. - PubMed
-
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

