Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas
- PMID: 20124221
- PMCID: PMC2877973
- DOI: 10.1074/mcp.M900398-MCP200
Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas
Abstract
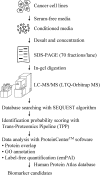
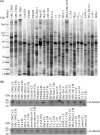
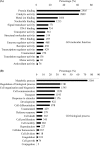
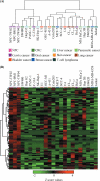
Although cancer cell secretome profiling is a promising strategy used to identify potential body fluid-accessible cancer biomarkers, questions remain regarding the depth to which the cancer cell secretome can be mined and the efficiency with which researchers can select useful candidates from the growing list of identified proteins. Therefore, we analyzed the secretomes of 23 human cancer cell lines derived from 11 cancer types using one-dimensional SDS-PAGE and nano-LC-MS/MS performed on an LTQ-Orbitrap mass spectrometer to generate a more comprehensive cancer cell secretome. A total of 31,180 proteins was detected, accounting for 4,584 non-redundant proteins, with an average of 1,300 proteins identified per cell line. Using protein secretion-predictive algorithms, 55.8% of the proteins appeared to be released or shed from cells. The identified proteins were selected as potential marker candidates according to three strategies: (i) proteins apparently secreted by one cancer type but not by others (cancer type-specific marker candidates), (ii) proteins released by most cancer cell lines (pan-cancer marker candidates), and (iii) proteins putatively linked to cancer-relevant pathways. We then examined protein expression profiles in the Human Protein Atlas to identify biomarker candidates that were simultaneously detected in the secretomes and highly expressed in cancer tissues. This analysis yielded 6-137 marker candidates selective for each tumor type and 94 potential pan-cancer markers. Among these, we selectively validated monocyte differentiation antigen CD14 (for liver cancer), stromal cell-derived factor 1 (for lung cancer), and cathepsin L1 and interferon-induced 17-kDa protein (for nasopharyngeal carcinoma) as potential serological cancer markers. In summary, the proteins identified from the secretomes of 23 cancer cell lines and the Human Protein Atlas represent a focused reservoir of potential cancer biomarkers.
Figures

References
-
- Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) Cancer statistics, 2008. CA Cancer J. Clin 58, 71–96 - PubMed
-
- Bettendorf O., Piffkò J., Bànkfalvi A. (2004) Prognostic and predictive factors in oral squamous cell cancer: important tools for planning individual therapy? Oral. Oncol 40, 110–119 - PubMed
-
- Conrads T. P., Zhou M., Petricoin E. F., 3rd, Liotta L., Veenstra T. D. (2003) Cancer diagnosis using proteomic patterns. Expert Rev. Mol. Diagn 3, 411–420 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

