The endosymbiotic origin, diversification and fate of plastids
- PMID: 20124341
- PMCID: PMC2817223
- DOI: 10.1098/rstb.2009.0103
The endosymbiotic origin, diversification and fate of plastids
Abstract
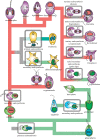
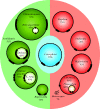
Plastids and mitochondria each arose from a single endosymbiotic event and share many similarities in how they were reduced and integrated with their host. However, the subsequent evolution of the two organelles could hardly be more different: mitochondria are a stable fixture of eukaryotic cells that are neither lost nor shuffled between lineages, whereas plastid evolution has been a complex mix of movement, loss and replacement. Molecular data from the past decade have substantially untangled this complex history, and we now know that plastids are derived from a single endosymbiotic event in the ancestor of glaucophytes, red algae and green algae (including plants). The plastids of both red algae and green algae were subsequently transferred to other lineages by secondary endosymbiosis. Green algal plastids were taken up by euglenids and chlorarachniophytes, as well as one small group of dinoflagellates. Red algae appear to have been taken up only once, giving rise to a diverse group called chromalveolates. Additional layers of complexity come from plastid loss, which has happened at least once and probably many times, and replacement. Plastid loss is difficult to prove, and cryptic, non-photosynthetic plastids are being found in many non-photosynthetic lineages. In other cases, photosynthetic lineages are now understood to have evolved from ancestors with a plastid of different origin, so an ancestral plastid has been replaced with a new one. Such replacement has taken place in several dinoflagellates (by tertiary endosymbiosis with other chromalveolates or serial secondary endosymbiosis with a green alga), and apparently also in two rhizarian lineages: chlorarachniophytes and Paulinella (which appear to have evolved from chromalveolate ancestors). The many twists and turns of plastid evolution each represent major evolutionary transitions, and each offers a glimpse into how genomes evolve and how cells integrate through gene transfers and protein trafficking.
Figures

References
-
- Abrahamsen M. S., et al. 2004Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445 (doi:10.1126/science.1094786) - DOI - PubMed
-
- Adl S. M., et al. 2005The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 (doi:10.1111/j.1550-7408.2005.00053.x) - DOI - PubMed
-
- Archibald J. M.2005Jumping genes and shrinking genomes—probing the evolution of eukaryotic photosynthesis with genomics. IUBMB Life 57, 539–547 (doi:10.1080/15216540500167732) - DOI - PubMed
-
- Archibald J. M.2007Nucleomorph genomes: structure, function, origin and evolution. Bioessays 29, 392–402 (doi:10.1002/bies.20551) - DOI - PubMed
-
- Archibald J. M., Keeling P. J.2002Recycled plastids: a green movement in eukaryotic evolution. Trends Genet. 18, 577–584 (doi:10.1016/S0168-9525(02)02777-4) - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
