Peroxisome diversity and evolution
- PMID: 20124343
- PMCID: PMC2817229
- DOI: 10.1098/rstb.2009.0240
Peroxisome diversity and evolution
Abstract
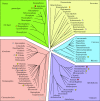
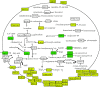
Peroxisomes are organelles bounded by a single membrane that can be found in all major groups of eukaryotes. A single evolutionary origin of this cellular compartment is supported by the presence, in diverse organisms, of a common set of proteins implicated in peroxisome biogenesis and maintenance. Their enzymatic content, however, can vary substantially across species, indicating a high level of evolutionary plasticity. Proteomic analyses have greatly expanded our knowledge on peroxisomes in some model organisms, including plants, mammals and yeasts. However, we still have a limited knowledge about the distribution and functionalities of peroxisomes in the vast majority of groups of microbial eukaryotes. Here, I review recent advances in our understanding of peroxisome diversity and evolution, with a special emphasis on peroxisomes in microbial eukaryotes.
Figures


Similar articles
-
The evolutionary origin of peroxisomes: an ER-peroxisome connection.Mol Biol Evol. 2006 Apr;23(4):838-45. doi: 10.1093/molbev/msj103. Epub 2006 Feb 1. Mol Biol Evol. 2006. PMID: 16452116
-
Evolution of the Peroxisomal Proteome.Subcell Biochem. 2018;89:221-233. doi: 10.1007/978-981-13-2233-4_9. Subcell Biochem. 2018. PMID: 30378025 Review.
-
Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):799-817. doi: 10.1098/rstb.2009.0167. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124346 Free PMC article. Review.
-
Yeast peroxisomes: How are they formed and how do they grow?Int J Biochem Cell Biol. 2018 Dec;105:24-34. doi: 10.1016/j.biocel.2018.09.019. Epub 2018 Sep 27. Int J Biochem Cell Biol. 2018. PMID: 30268746 Review.
-
Peroxisomes: offshoots of the ER.Curr Opin Cell Biol. 2013 Aug;25(4):449-54. doi: 10.1016/j.ceb.2013.05.004. Epub 2013 Jun 14. Curr Opin Cell Biol. 2013. PMID: 23773570 Review.
Cited by
-
Oxidant sensing by reversible disulfide bond formation.J Biol Chem. 2013 Sep 13;288(37):26489-96. doi: 10.1074/jbc.R113.462929. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861395 Free PMC article. Review.
-
Import of proteins into the peroxisomal matrix.Front Physiol. 2013 Sep 24;4:261. doi: 10.3389/fphys.2013.00261. Front Physiol. 2013. PMID: 24069002 Free PMC article. Review.
-
Peroxisome proliferator activated receptors at the crossroad of obesity, diabetes, and pancreatic cancer.World J Gastroenterol. 2016 Feb 28;22(8):2441-59. doi: 10.3748/wjg.v22.i8.2441. World J Gastroenterol. 2016. PMID: 26937133 Free PMC article. Review.
-
Melatonin-Nrf2 Signaling Activates Peroxisomal Activities in Porcine Cumulus Cell-Oocyte Complexes.Antioxidants (Basel). 2020 Nov 3;9(11):1080. doi: 10.3390/antiox9111080. Antioxidants (Basel). 2020. PMID: 33153240 Free PMC article.
-
Origin and evolution of metabolic sub-cellular compartmentalization in eukaryotes.Biochimie. 2015 Dec;119:262-8. doi: 10.1016/j.biochi.2015.03.021. Epub 2015 Apr 11. Biochimie. 2015. PMID: 25869000 Free PMC article. Review.
References
-
- Agne B., Meindl N. M., Niederhoff K., Einwachter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., Kunau W. H.2003Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646 (doi:10.1016/S1097-2765(03)00062-5) - DOI - PubMed
-
- Armbrust E. V., et al. 2004The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 (doi:10.1126/science.1101156) - DOI - PubMed
-
- Bernhard J. M., Bowser S. S.2008Peroxisome proliferation in Foraminifera inhabiting the chemocline: an adaptation to reactive oxygen species exposure? J. Eukaryot. Microbiol. 55, 135–144 (doi:10.1111/j.1550-7408.2008.00318.x) - DOI - PMC - PubMed
-
- Birdsey G. M., Lewin J., Cunningham A. A., Bruford M. W., Danpure C. J.2004Differential enzyme targeting as an evolutionary adaptation to herbivory in carnivora. Mol. Biol. Evol. 21, 632–646 (doi:10.1093/molbev/msh054) - DOI - PubMed
-
- Brocard C., Hartig A.2006Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

