Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes
- PMID: 20124346
- PMCID: PMC2817224
- DOI: 10.1098/rstb.2009.0167
Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes
Abstract
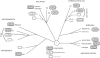
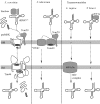
All eukaryotes require mitochondria for survival and growth. The origin of mitochondria can be traced down to a single endosymbiotic event between two probably prokaryotic organisms. Subsequent evolution has left mitochondria a collection of heterogeneous organelle variants. Most of these variants have retained their own genome and translation system. In hydrogenosomes and mitosomes, however, the entire genome was lost. All types of mitochondria import most of their proteome from the cytosol, irrespective of whether they have a genome or not. Moreover, in most eukaryotes, a variable number of tRNAs that are required for mitochondrial translation are also imported. Thus, import of macromolecules, both proteins and tRNA, is essential for mitochondrial biogenesis. Here, we review what is known about the evolutionary history of the two processes using a recently revised eukaryotic phylogeny as a framework. We discuss how the processes of protein import and tRNA import relate to each other in an evolutionary context.
Figures



References
-
- Adams K. L., Palmer J. D.2003Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395 (doi:10.1016/S1055-7903(03)00194-5) - DOI - PubMed
-
- Adhya S.2008Leishmania mitochondrial tRNA importers. Int. J. Biochem. Cell. Biol. 12, 2681–2685 (doi:10.1016/j.biocel.2007.10.025) - DOI - PubMed
-
- Adl S. M., et al. 2005The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 (doi:10.1111/j.1550-7408.2005.00053.x) - DOI - PubMed
-
- Aguilera P., Barry T., Tovar J.2008Entamoeba histolytica mitosomes: organelles in search of a function. Exp. Parasitol. 118, 10–16 (doi:10.1016/j.exppara.2007.08.004) - DOI - PubMed
-
- Alfonzo J. D., Blanc V., Estevez A. M., Rubio M. A. T., Simpson L.1999C to U editing of anticodon of imported mitochondrial tRNATrp allows decoding of UGA stop codon in Leishmania. EMBO J. 18, 7056–7062 (doi:10.1093/emboj/18.24.7056) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
