A humanized UGT1 mouse model expressing the UGT1A1*28 allele for assessing drug clearance by UGT1A1-dependent glucuronidation
- PMID: 20124398
- PMCID: PMC2872941
- DOI: 10.1124/dmd.109.030130
A humanized UGT1 mouse model expressing the UGT1A1*28 allele for assessing drug clearance by UGT1A1-dependent glucuronidation
Abstract
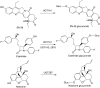
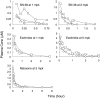
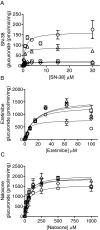
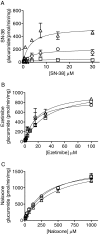
Humanized mice that express the human UDP-glucuronosyltransferase (UGT) 1 locus have been developed in a Ugt1-null background as a model to improve predictions of human UGT1A-dependent drug clearance. Enzyme kinetic parameters (K(m) and V(max)) and pharmacokinetic properties of three probe drugs were compared using wild-type and humanized UGT1 mice that express the Gilbert's UGT1A1*28 allele [Tg(UGT1(A1*28)) Ugt1(-/-) mice]. The well characterized substrate for UGT1A1, 7-ethyl-10-hydroxy-camptothecin (SN-38), showed the greatest difference in parent drug exposure ( approximately 3-fold increase) and clearance ( approximately 3-fold decrease) in Tg(UGT1(A1*28)) Ugt1(-/-) mice after intravenous administration compared with wild-type and phenobarbital-treated animals. In contrast, the clearance of the UGT2B7 substrate (-)-17-allyl-4, 5alpha-epoxy-3, 14-dihydroxymorphinan-6-one (naloxone) was not altered in Tg(UGT1(A1*28)) Ugt1(-/-) mice. In addition, pharmacokinetic parameters with 1-(4-fluorophenyl)3(R)-[3-(4-fluorophenyl)-3(S)-hydroxypropyl]-4(S)-(4-hydroxyphenyl)-2-azetidinone (ezetimibe, Zetia; Merck & Co., Whitehouse Station, NJ), considered to be a major substrate for UGT1A1, showed small to no dependence on UGT1A1-directed glucuronidation. Enzyme kinetic parameters assessed for SN-38, ezetimibe, and naloxone using liver microsomes prepared from wild-type and Tg(UGT1(A1*28)) Ugt1(-/-) mice showed patterns consistent with the in vivo pharmacokinetic data. For SN-38 glucuronidation, V(max) decreased 5-fold in Tg(UGT1(A1*28)) Ugt1(-/-) mouse liver microsomes compared with microsomes prepared from wild-type mice, and decreased 10-fold compared with phenobarbital-treated Tg(UGT1(A1*28)) Ugt1(-/-) mice. These differences are consistent with SN-38 glucuronidation activities using HLMs isolated from individuals genotyped as UGT1A1*1 or UGT1A1*28. For ezetimibe and naloxone the differences in V(max) were minimal. Thus, Tg(UGT1(A1*28)) Ugt1(-/-) mice can serve as a pharmacokinetic model to further investigate the effects of UGT1A1 expression on drug metabolism.
Figures
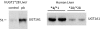

Similar articles
-
Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia).Drug Metab Dispos. 2004 Mar;32(3):314-20. doi: 10.1124/dmd.32.3.314. Drug Metab Dispos. 2004. PMID: 14977865
-
Intestinal glucuronidation protects against chemotherapy-induced toxicity by irinotecan (CPT-11).Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):19143-8. doi: 10.1073/pnas.1319123110. Epub 2013 Nov 4. Proc Natl Acad Sci U S A. 2013. PMID: 24191041 Free PMC article.
-
Developmental hyperbilirubinemia and CNS toxicity in mice humanized with the UDP glucuronosyltransferase 1 (UGT1) locus.Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):5024-9. doi: 10.1073/pnas.0913290107. Epub 2010 Mar 1. Proc Natl Acad Sci U S A. 2010. PMID: 20194756 Free PMC article.
-
Species differences in drug glucuronidation: Humanized UDP-glucuronosyltransferase 1 mice and their application for predicting drug glucuronidation and drug-induced toxicity in humans.Drug Metab Pharmacokinet. 2018 Feb;33(1):9-16. doi: 10.1016/j.dmpk.2017.10.002. Epub 2017 Oct 7. Drug Metab Pharmacokinet. 2018. PMID: 29079228 Free PMC article. Review.
-
Pharmacogenetics of uridine diphosphoglucuronosyltransferase (UGT) 1A family members and its role in patient response to irinotecan.Drug Metab Rev. 2006;38(3):393-409. doi: 10.1080/03602530600739835. Drug Metab Rev. 2006. PMID: 16877259 Review.
Cited by
-
Glucuronidation of drugs in humanized UDP-glucuronosyltransferase 1 mice: Similarity with glucuronidation in human liver microsomes.Pharmacol Res Perspect. 2013 Oct;1(1):e00002. doi: 10.1002/prp2.2. Epub 2013 Sep 3. Pharmacol Res Perspect. 2013. PMID: 25505556 Free PMC article.
-
Drug-metabolizing enzyme, transporter, and nuclear receptor genetically modified mouse models.Drug Metab Rev. 2011 Feb;43(1):27-40. doi: 10.3109/03602532.2010.512294. Epub 2010 Sep 21. Drug Metab Rev. 2011. PMID: 20854191 Free PMC article. Review.
-
Humanized transgenic mouse models for drug metabolism and pharmacokinetic research.Curr Drug Metab. 2011 Dec;12(10):997-1006. doi: 10.2174/138920011798062265. Curr Drug Metab. 2011. PMID: 22023319 Free PMC article.
-
Altered gene expression profiles in the lungs of benzo[a]pyrene-exposed mice in the presence of lipopolysaccharide-induced pulmonary inflammation.Toxicol Appl Pharmacol. 2017 Dec 1;336:8-19. doi: 10.1016/j.taap.2017.09.023. Epub 2017 Oct 5. Toxicol Appl Pharmacol. 2017. PMID: 28987381 Free PMC article.
-
Xenobiotic metabolomics: major impact on the metabolome.Annu Rev Pharmacol Toxicol. 2012;52:37-56. doi: 10.1146/annurev-pharmtox-010611-134748. Epub 2011 Aug 3. Annu Rev Pharmacol Toxicol. 2012. PMID: 21819238 Free PMC article. Review.
References
-
- Ando Y, Fujita K, Sasaki Y, Hasegawa Y. (2007) UGT1AI*6 and UGT1A1*27 for individualized irinotecan chemotherapy. Curr Opin Mol Ther 9:258–262 - PubMed
-
- Beaumont K, Smith DA. (2009) Does human pharmacokinetic prediction add significant value to compound selection in drug discovery research? Curr Opin Drug Discov Devel 12:61–71 - PubMed
-
- Bosma PJ. (2003) Inherited disorders of bilirubin metabolism. J Hepatol 38:107–117 - PubMed
-
- Caldwell GW, Masucci JA, Yan Z, Hageman W. (2004) Allometric scaling of pharmacokinetic parameters in drug discovery: can human CL, Vss and t1/2 be predicted from in-vivo rat data? Eur J Drug Metab Pharmacokinet 29:133–143 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

