Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion
- PMID: 20124417
- PMCID: PMC2823575
- DOI: 10.1242/jcs.057984
Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion
Abstract
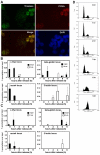
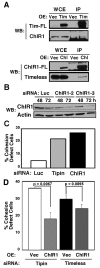
The Timeless-Tipin protein complex has been reported to be important for replication checkpoint and normal DNA replication processes. However, the precise mechanisms by which Timeless-Tipin preserves genomic integrity are largely unclear. Here, we describe the roles of Timeless-Tipin in replication fork stabilization and sister chromatid cohesion. We show in human cells that Timeless is recruited to replication origin regions and dissociate from them as replication proceeds. Cdc45, which is known to be required for replication fork progression, shows similar patterns of origin association to those of Timeless. Depletion of Timeless-Tipin causes chromosome fragmentation and defects in damage repair in response to fork collapse, suggesting that it is required for replication fork maintenance under stress. We also demonstrate that depletion of Timeless-Tipin impairs sister chromatid cohesion and causes a defect in mitotic progression. Consistently, Timeless-Tipin co-purifies with cohesin subunits and is required for their stable association with chromatin during S phase. Timeless associates with the cohesion-promoting DNA helicase ChlR1, which, when overexpressed, partially alleviates the cohesion defect of cells depleted of Timeless-Tipin. These results suggest that Timeless-Tipin functions as a replication fork stabilizer that couples DNA replication with sister chromatid cohesion established at replication forks.
Figures





References
-
- Aguilera A., Gomez-Gonzalez B. (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204-217 - PubMed
-
- Ansbach A. B., Noguchi C., Klansek I. W., Heidlebaugh M., Nakamura T. M., Noguchi E. (2008). RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol. Biol. Cell 19, 595-607 - PMC - PubMed
-
- Barnes J. W., Tischkau S. A., Barnes J. A., Mitchell J. W., Burgoon P. W., Hickok J. R., Gillette M. U. (2003). Requirement of mammalian Timeless for circadian rhythmicity. Science 302, 439-442 - PubMed
-
- Bartek J., Lukas J. (2007). DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238-245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

