Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI 60 cancer cell line set
- PMID: 20124458
- PMCID: PMC4085051
- DOI: 10.1158/1535-7163.MCT-09-0743
Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI 60 cancer cell line set
Abstract
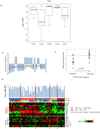
Aberrations in oncogenes and tumor suppressors frequently affect the activity of critical signal transduction pathways. To analyze systematically the relationship between the activation status of protein networks and other characteristics of cancer cells, we did reverse phase protein array (RPPA) profiling of the NCI60 cell lines for total protein expression and activation-specific markers of critical signaling pathways. To extend the scope of the study, we merged those data with previously published RPPA results for the NCI60. Integrative analysis of the expanded RPPA data set revealed five major clusters of cell lines and five principal proteomic signatures. Comparison of mutations in the NCI60 cell lines with patterns of protein expression showed significant associations for PTEN, PIK3CA, BRAF, and APC mutations with proteomic clusters. PIK3CA and PTEN mutation enrichment were not cell lineage-specific but were associated with dominant yet distinct groups of proteins. The five RPPA-defined clusters were strongly associated with sensitivity to standard anticancer agents. RPPA analysis identified 27 protein features significantly associated with sensitivity to paclitaxel. The functional status of those proteins was interrogated in a paclitaxel whole genome small interfering RNA (siRNA) library synthetic lethality screen and confirmed the predicted associations with drug sensitivity. These studies expand our understanding of the activation status of protein networks in the NCI60 cancer cell lines, demonstrate the importance of the direct study of protein expression and activation, and provide a basis for further studies integrating the information with other molecular and pharmacological characteristics of cancer.
Figures


References
-
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. - PubMed
-
- Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–1491. - PubMed
-
- Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

