Crystallization and preliminary X-ray diffraction analyses of the homodimeric glycine decarboxylase (P-protein) from the cyanobacterium Synechocystis sp. PCC 6803
- PMID: 20124719
- PMCID: PMC2815689
- DOI: 10.1107/S1744309109052828
Crystallization and preliminary X-ray diffraction analyses of the homodimeric glycine decarboxylase (P-protein) from the cyanobacterium Synechocystis sp. PCC 6803
Abstract
Glycine decarboxylase, or P-protein, is a major enzyme that is involved in the C(1) metabolism of all organisms and in the photorespiratory pathway of plants and cyanobacteria. The protein from Synechocystis sp. PCC 6803 is a homodimer with a mass of 215 kDa. Recombinant glycine decarboxylase was expressed in Escherichia coli and purified by metal-affinity, ion-exchange and gel-filtration chromatography. Crystals of P-protein that diffracted to a resolution of 2.1 A were obtained using the hanging-drop vapour-diffusion method at 291 K. X-ray diffraction data were collected from cryocooled crystals using synchrotron radiation. The crystals belonged to space group P2(1)2(1)2(1), with unit-cell parameters a = 96.30, b = 135.81, c = 179.08 A.
Figures

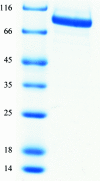

Similar articles
-
Structure of the homodimeric glycine decarboxylase P-protein from Synechocystis sp. PCC 6803 suggests a mechanism for redox regulation.J Biol Chem. 2013 Dec 6;288(49):35333-45. doi: 10.1074/jbc.M113.509976. Epub 2013 Oct 11. J Biol Chem. 2013. PMID: 24121504 Free PMC article.
-
Properties of recombinant glycine decarboxylase P- and H-protein subunits from the cyanobacterium Synechocystis sp. strain PCC 6803.FEBS Lett. 2007 Apr 3;581(7):1297-301. doi: 10.1016/j.febslet.2007.02.037. Epub 2007 Feb 28. FEBS Lett. 2007. PMID: 17349627
-
Purification, crystallization and preliminary X-ray diffraction analysis of ArsH from Synechocystis sp. strain PCC 6803.Acta Crystallogr F Struct Biol Commun. 2014 Apr;70(Pt 4):497-500. doi: 10.1107/S2053230X14004865. Epub 2014 Mar 25. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24699748 Free PMC article.
-
Cloning, purification, crystallization and preliminary X-ray crystallographic analysis of MCAT from Synechocystis sp. PCC 6803.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Nov;69(Pt 11):1256-9. doi: 10.1107/S1744309113026274. Epub 2013 Oct 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 24192363 Free PMC article.
-
Coexpression, purification, crystallization and preliminary X-ray characterization of glycine decarboxylase (P-protein) of the glycine-cleavage system from Thermus thermophilus HB8.Acta Crystallogr D Biol Crystallogr. 2003 Mar;59(Pt 3):554-7. doi: 10.1107/s090744490300043x. Epub 2003 Feb 21. Acta Crystallogr D Biol Crystallogr. 2003. PMID: 12595724
Cited by
-
Structure of the homodimeric glycine decarboxylase P-protein from Synechocystis sp. PCC 6803 suggests a mechanism for redox regulation.J Biol Chem. 2013 Dec 6;288(49):35333-45. doi: 10.1074/jbc.M113.509976. Epub 2013 Oct 11. J Biol Chem. 2013. PMID: 24121504 Free PMC article.
-
Phycocompounds from Cyanobacteria: Exploring Synergistic Effects with Conventional Anticancer and Antimicrobial Properties.ACS Omega. 2025 Jun 5;10(23):23957-23980. doi: 10.1021/acsomega.5c03526. eCollection 2025 Jun 17. ACS Omega. 2025. PMID: 40547631 Free PMC article. Review.
References
-
- Douce, R., Bourguignon, J., Neuburger, M. & Rebeille, F. (2001). Trends Plant Sci.6, 167–176. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

