Rare variants create synthetic genome-wide associations
- PMID: 20126254
- PMCID: PMC2811148
- DOI: 10.1371/journal.pbio.1000294
Rare variants create synthetic genome-wide associations
Abstract
Genome-wide association studies (GWAS) have now identified at least 2,000 common variants that appear associated with common diseases or related traits (http://www.genome.gov/gwastudies), hundreds of which have been convincingly replicated. It is generally thought that the associated markers reflect the effect of a nearby common (minor allele frequency >0.05) causal site, which is associated with the marker, leading to extensive resequencing efforts to find causal sites. We propose as an alternative explanation that variants much less common than the associated one may create "synthetic associations" by occurring, stochastically, more often in association with one of the alleles at the common site versus the other allele. Although synthetic associations are an obvious theoretical possibility, they have never been systematically explored as a possible explanation for GWAS findings. Here, we use simple computer simulations to show the conditions under which such synthetic associations will arise and how they may be recognized. We show that they are not only possible, but inevitable, and that under simple but reasonable genetic models, they are likely to account for or contribute to many of the recently identified signals reported in genome-wide association studies. We also illustrate the behavior of synthetic associations in real datasets by showing that rare causal mutations responsible for both hearing loss and sickle cell anemia create genome-wide significant synthetic associations, in the latter case extending over a 2.5-Mb interval encompassing scores of "blocks" of associated variants. In conclusion, uncommon or rare genetic variants can easily create synthetic associations that are credited to common variants, and this possibility requires careful consideration in the interpretation and follow up of GWAS signals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
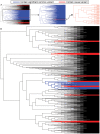
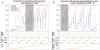
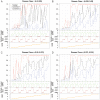

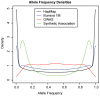


Comment in
-
Synthetic associations created by rare variants do not explain most GWAS results.PLoS Biol. 2011 Jan 18;9(1):e1000579. doi: 10.1371/journal.pbio.1000579. PLoS Biol. 2011. PMID: 21267061 Free PMC article. No abstract available.
-
Synthetic associations are unlikely to account for many common disease genome-wide association signals.PLoS Biol. 2011 Jan 18;9(1):e1000580. doi: 10.1371/journal.pbio.1000580. PLoS Biol. 2011. PMID: 21267062 Free PMC article.
-
Rare treasures.Nat Rev Genet. 2010 Mar;11(3):170. doi: 10.1038/nrg2754. Nat Rev Genet. 2010. PMID: 21485427 No abstract available.
References
-
- Lowe C. E, Cooper J. D, Brusko T, Walker N. M, Smyth D. J, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. - PubMed
-
- Burfoot R. K, Jensen C. J, Field J, Stankovich J, Varney M. D, et al. SNP mapping and candidate gene sequencing in the class I region of the HLA complex: searching for multiple sclerosis susceptibility genes in Tasmanians. Tissue Antigens. 2008;71:42–50. - PubMed
-
- Deloukas P on behalf of the Wellcome Trust Case Control Consortium. High throughput approaches to fine mapping in regions of confirmed association. 2008. Presentation at the 58th Annual Meeting of the American Society of Human Genetics, November 13, 2008; Philadelphia, Pennsylvania.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

