SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas
- PMID: 20126410
- PMCID: PMC2813300
- DOI: 10.1371/journal.pone.0008960
SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas
Abstract
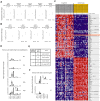
Squamous cell carcinoma (SCC) of the lung is a frequent and aggressive cancer type. Gene amplifications, a known activating mechanism of oncogenes, target the 3q26-qter region as one of the most frequently gained/amplified genomic sites in SCC of various types. Here, we used array comparative genomic hybridization to delineate the consensus region of 3q26.3 amplifications in lung SCC. Recurrent amplifications occur in 20% of lung SCC (136 tumors in total) and map to a core region of 2 Mb (Megabases) that encompasses SOX2, a transcription factor gene. Intense SOX2 immunostaining is frequent in nuclei of lung SCC, indicating potential active transcriptional regulation by SOX2. Analyses of the transcriptome of lung SCC, SOX2-overexpressing lung epithelial cells and embryonic stem cells (ESCs) reveal that SOX2 contributes to activate ESC-like phenotypes and provide clues pertaining to the deregulated genes involved in the malignant phenotype. In cell culture experiments, overexpression of SOX2 stimulates cellular migration and anchorage-independent growth while SOX2 knockdown impairs cell growth. Finally, SOX2 over-expression in non-tumorigenic human lung bronchial epithelial cells is tumorigenic in immunocompromised mice. These results indicate that the SOX2 transcription factor, a major regulator of stem cell function, is also an oncogene and a driver gene for the recurrent 3q26.33 amplifications in lung SCC.
Conflict of interest statement
Figures




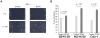


References
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
-
- Hogarty MD, Brodeur GM. Scriver CR, Sly WS, Childs B, Beaudet AL, Valle D, Kinzler KW, Vogelstein B, editors. Gene Amplification in Human Cancers: Biological and Clinical Significance. The Metabolic & Molecular Bases of Inherited Disease, 8th edition. 2001. pp. 597–610.
-
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. - PubMed
-
- Gebhart E, Liehr T. Patterns of genomic imbalances in human solid tumors. Int J Oncol. 2000;162:383–399. - PubMed
-
- Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877–6883. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

