Genetics and genomics of melanoma
- PMID: 20126509
- PMCID: PMC2771951
- DOI: 10.1586/edm.09.2
Genetics and genomics of melanoma
Abstract
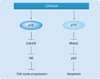
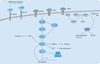
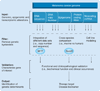
The rapidly increasing incidence of melanoma, coupled with its highly aggressive metastatic nature, is of urgent concern. In order to design rational therapies, it is of critical importance to identify the genetic determinants that drive melanoma formation and progression. To date, signaling cascades emanating from the EGF receptor, c-MET and other receptors are known to be altered in melanoma. Important mutations in signaling molecules, such as BRAF and N-RAS, have been identified. In this review, some of the major genetic alterations and signaling pathways involved in melanoma will be discussed. Given the great deal of genetic heterogeneity observed in melanoma, it is likely that many more genetic determinants exist. Through the use of powerful genomic technologies, it is now possible to identify these additional genetic alterations in melanoma. A critical step in this analysis will be culling bystanders from functionally important drivers, as this will highlight genetic elements that will be promising therapeutic targets. Such technologies and the important points to consider in understanding the genetics of melanoma will be reviewed.
Figures
Similar articles
-
Genetic progression of malignant melanoma.Cancer Metastasis Rev. 2016 Mar;35(1):93-107. doi: 10.1007/s10555-016-9613-5. Cancer Metastasis Rev. 2016. PMID: 26970965 Review.
-
[Characterization of genetic alterations in primary human melanomas carrying BRAF or NRAS mutation].Magy Onkol. 2013 Jun;57(2):96-9. Epub 2013 May 20. Magy Onkol. 2013. PMID: 23795354 Hungarian.
-
Genomic and Transcriptomic Underpinnings of Melanoma Genesis, Progression, and Metastasis.Cancers (Basel). 2021 Dec 28;14(1):123. doi: 10.3390/cancers14010123. Cancers (Basel). 2021. PMID: 35008286 Free PMC article. Review.
-
Genomic aberrations in spitzoid melanocytic tumours and their implications for diagnosis, prognosis and therapy.Pathology. 2016 Feb;48(2):113-31. doi: 10.1016/j.pathol.2015.12.007. Epub 2016 Jan 18. Pathology. 2016. PMID: 27020384 Free PMC article. Review.
-
Targeting RAS/RAF/MEK/ERK signaling in metastatic melanoma.IUBMB Life. 2013 Sep;65(9):748-58. doi: 10.1002/iub.1193. Epub 2013 Jul 29. IUBMB Life. 2013. PMID: 23893853 Review.
Cited by
-
Deep-proteome mapping of WM-266-4 human metastatic melanoma cells: From oncogenic addiction to druggable targets.PLoS One. 2017 Feb 3;12(2):e0171512. doi: 10.1371/journal.pone.0171512. eCollection 2017. PLoS One. 2017. PMID: 28158294 Free PMC article.
-
Targeting Glutamatergic Signaling and the PI3 Kinase Pathway to Halt Melanoma Progression.Transl Oncol. 2015 Feb;8(1):1-9. doi: 10.1016/j.tranon.2014.11.001. Transl Oncol. 2015. PMID: 25749171 Free PMC article.
-
Sensitivity of Melanoma Cells to EGFR and FGFR Activation but Not Inhibition is Influenced by Oncogenic BRAF and NRAS Mutations.Pathol Oncol Res. 2015 Sep;21(4):957-68. doi: 10.1007/s12253-015-9916-9. Epub 2015 Mar 9. Pathol Oncol Res. 2015. PMID: 25749811
-
Aggressiveness of human melanoma xenograft models is promoted by aneuploidy-driven gene expression deregulation.Oncotarget. 2012 Apr;3(4):399-413. doi: 10.18632/oncotarget.473. Oncotarget. 2012. PMID: 22535842 Free PMC article.
-
Bioinformatic and Machine Learning Applications in Melanoma Risk Assessment and Prognosis: A Literature Review.Genes (Basel). 2021 Oct 30;12(11):1751. doi: 10.3390/genes12111751. Genes (Basel). 2021. PMID: 34828357 Free PMC article. Review.
References
-
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–857. - PubMed
-
- Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, editors. National Cancer Institute. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD, USA: National Cancer Institute; 2008.
-
- Rigel DS. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J. Am. Acad. Dermatol. 2008;58(5 Suppl 2):S129–S132. - PubMed
-
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat. Rev. Cancer. 2003;3(8):559–570. - PubMed
-
- Swetter SM, Boldrick JC, Jung SY, Egbert BM, Harvell JD. Increasing incidence of lentigo maligna melanoma subtypes: northern California and national trends 1990–2000. J. Invest. Dermatol. 2005;125(4):685–691. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
