Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System
- PMID: 20126518
- PMCID: PMC2805431
- DOI: 10.3389/neuro.04.021.2009
Optogenetic Dissection of Neuronal Circuits in Zebrafish using Viral Gene Transfer and the Tet System
Abstract
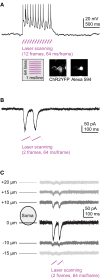
The conditional expression of transgenes at high levels in sparse and specific populations of neurons is important for high-resolution optogenetic analyses of neuronal circuits. We explored two complementary methods, viral gene delivery and the iTet-Off system, to express transgenes in the brain of zebrafish. High-level gene expression in neurons was achieved by Sindbis and Rabies viruses. The Tet system produced strong and specific gene expression that could be modulated conveniently by doxycycline. Moreover, transgenic lines showed expression in distinct, sparse and stable populations of neurons that appeared to be subsets of the neurons targeted by the promoter driving the Tet-activator. The Tet system therefore provides the opportunity to generate libraries of diverse expression patterns similar to gene trap approaches or the thy-1 promoter in mice, but with the additional possibility to pre-select cell types of interest. In transgenic lines expressing channelrhodopsin-2, action potential firing could be precisely controlled by two-photon stimulation at low laser power, presumably because the expression levels of the Tet-controlled genes were high even in adults. In channelrhodopsin-2-expressing larvae, optical stimulation with a single blue LED evoked distinct swimming behaviors including backward swimming. These approaches provide new opportunities for the optogenetic dissection of neuronal circuit structure and function.
Keywords: Tet system; channelrhodopsin; multiphoton; olfactory bulb; optogenetics; viral gene transfer; zebrafish.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

