E. coli K-12 and EHEC genes regulated by SdiA
- PMID: 20126629
- PMCID: PMC2812512
- DOI: 10.1371/journal.pone.0008946
E. coli K-12 and EHEC genes regulated by SdiA
Abstract
Background: Escherichia and Salmonella encode SdiA, a transcription factor of the LuxR family that regulates genes in response to N-acyl homoserine lactones (AHLs) produced by other species of bacteria. E. coli genes that change expression in the presence of plasmid-encoded sdiA have been identified by several labs. However, many of these genes were identified by overexpressing sdiA on a plasmid and have not been tested for a response to sdiA produced from its natural position in the chromosome or for a response to AHL.
Methodology/principal findings: We determined that two important loci reported to respond to plasmid-based sdiA, ftsQAZ and acrAB, do not respond to sdiA expressed from its natural position in the chromosome or to AHLs. To identify genes that are regulated by chromosomal sdiA and/or AHLs, we screened 10,000 random transposon-based luciferase fusions in E. coli K-12 and a further 10,000 in E. coli O157:H7 for a response to AHL and then tested these genes for sdiA-dependence. We found that genes encoding the glutamate-dependent acid resistance system are up-regulated, and fliE is down-regulated, by sdiA. Gene regulation by sdiA of E. coli is only partially dependent upon AHL.
Conclusions/significance: The genes of E. coli that respond to plasmid-based expression of sdiA are largely different than those that respond to chromosomal sdiA and/or AHL. This has significant implications for determining the true function of AHL detection by E. coli.
Conflict of interest statement
Figures

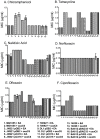
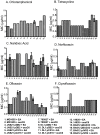
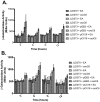
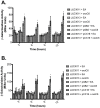





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

