The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing
- PMID: 20126651
- PMCID: PMC2811190
- DOI: 10.1371/journal.pone.0008888
The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing
Abstract
Background: We recently showed that enzymes of the TET family convert 5-mC to 5-hydroxymethylcytosine (5-hmC) in DNA. 5-hmC is present at high levels in embryonic stem cells and Purkinje neurons. The methylation status of cytosines is typically assessed by reaction with sodium bisulfite followed by PCR amplification. Reaction with sodium bisulfite promotes cytosine deamination, whereas 5-methylcytosine (5-mC) reacts poorly with bisulfite and is resistant to deamination. Since 5-hmC reacts with bisulfite to yield cytosine 5-methylenesulfonate (CMS), we asked how DNA containing 5-hmC behaves in bisulfite sequencing.
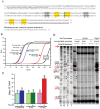
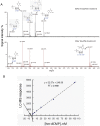
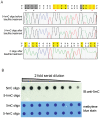
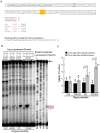
Methodology/principal findings: We used synthetic oligonucleotides with different distributions of cytosine as templates for generation of DNAs containing C, 5-mC and 5-hmC. The resulting DNAs were subjected in parallel to bisulfite treatment, followed by exposure to conditions promoting cytosine deamination. The extent of conversion of 5-hmC to CMS was estimated to be 99.7%. Sequencing of PCR products showed that neither 5-mC nor 5-hmC undergo C-to-T transitions after bisulfite treatment, confirming that these two modified cytosine species are indistinguishable by the bisulfite technique. DNA in which CMS constituted a large fraction of all bases (28/201) was much less efficiently amplified than DNA in which those bases were 5-mC or uracil (the latter produced by cytosine deamination). Using a series of primer extension experiments, we traced the inefficient amplification of CMS-containing DNA to stalling of Taq polymerase at sites of CMS modification, especially when two CMS bases were either adjacent to one another or separated by 1-2 nucleotides.
Conclusions: We have confirmed that the widely used bisulfite sequencing technique does not distinguish between 5-mC and 5-hmC. Moreover, we show that CMS, the product of bisulfite conversion of 5-hmC, tends to stall DNA polymerases during PCR, suggesting that densely hydroxymethylated regions of DNA may be underrepresented in quantitative methylation analyses.
Conflict of interest statement
Figures

Similar articles
-
High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue.J Vis Exp. 2011 Feb 1;(48):2661. doi: 10.3791/2661. J Vis Exp. 2011. PMID: 21307836 Free PMC article.
-
Does urea promote the bisulfite-mediated deamination of cytosine in DNA? Investigation aiming at speeding-up the procedure for DNA methylation analysis.Nucleic Acids Symp Ser (Oxf). 2006;(50):69-70. doi: 10.1093/nass/nrl034. Nucleic Acids Symp Ser (Oxf). 2006. PMID: 17150821
-
Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine.Nat Protoc. 2012 Dec;7(12):2159-70. doi: 10.1038/nprot.2012.137. Epub 2012 Nov 29. Nat Protoc. 2012. PMID: 23196972 Free PMC article.
-
The bisulfite genomic sequencing used in the analysis of epigenetic states, a technique in the emerging environmental genotoxicology research.Mutat Res. 2008 Jul-Aug;659(1-2):77-82. doi: 10.1016/j.mrrev.2008.04.003. Epub 2008 May 14. Mutat Res. 2008. PMID: 18485805 Review.
-
Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis--a personal account.Proc Jpn Acad Ser B Phys Biol Sci. 2008;84(8):321-30. doi: 10.2183/pjab.84.321. Proc Jpn Acad Ser B Phys Biol Sci. 2008. PMID: 18941305 Free PMC article. Review.
Cited by
-
Solid-state nanopore analysis of human genomic DNA shows unaltered global 5-hydroxymethylcytosine content associated with early-stage breast cancer.Nanomedicine. 2021 Jul;35:102407. doi: 10.1016/j.nano.2021.102407. Epub 2021 Apr 24. Nanomedicine. 2021. PMID: 33905828 Free PMC article.
-
Epi-fingerprinting and epi-interventions for improved crop production and food quality.Front Plant Sci. 2015 Jun 5;6:397. doi: 10.3389/fpls.2015.00397. eCollection 2015. Front Plant Sci. 2015. PMID: 26097484 Free PMC article. Review.
-
Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells.Cell Stem Cell. 2013 Apr 4;12(4):470-8. doi: 10.1016/j.stem.2013.01.016. Epub 2013 Feb 14. Cell Stem Cell. 2013. PMID: 23415914 Free PMC article.
-
Eyes on DNA methylation: current evidence for DNA methylation in ocular development and disease.J Ocul Biol Dis Infor. 2011 Sep;4(3):95-103. doi: 10.1007/s12177-012-9078-x. Epub 2012 Mar 29. J Ocul Biol Dis Infor. 2011. PMID: 23538551 Free PMC article.
-
A mechanistic role for DNA methylation in endothelial cell (EC)-enriched gene expression: relationship with DNA replication timing.Blood. 2013 Apr 25;121(17):3531-40. doi: 10.1182/blood-2013-01-479170. Epub 2013 Feb 28. Blood. 2013. PMID: 23449636 Free PMC article.
References
-
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. - PubMed
-
- Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. - PubMed
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. - PubMed
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

