Microfluidic isolation and transcriptome analysis of serum microvesicles
- PMID: 20126692
- PMCID: PMC3136803
- DOI: 10.1039/b916199f
Microfluidic isolation and transcriptome analysis of serum microvesicles
Abstract
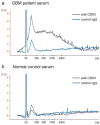
Microvesicles (exosomes) shed from both normal and cancerous cells may serve as means of intercellular communication. These microvesicles carry proteins, lipids and nucleic acids derived from the host cell. Their isolation and analysis from blood samples have the potential to provide information about state and progression of malignancy and should prove of great clinical importance as biomarkers for a variety of disease states. However, current protocols for isolation of microvesicles from blood require high-speed centrifugation and filtration, which are cumbersome and time consuming. In order to take full advantage of the potential of microvesicles as biomarkers for clinical applications, faster and simpler methods of isolation will be needed. In this paper, we present an easy and rapid microfluidic immunoaffinity method to isolate microvesicles from small volumes of both serum from blood samples and conditioned medium from cells in culture. RNA of high quality can be extracted from these microvesicles providing a source of information about the genetic status of tumors to serve as biomarkers for diagnosis and prognosis of cancer.
Figures



References
-
- Andre F, Schartz NEC, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Lancet. 2002;360:295–305. - PubMed
-
- Thery C, Zitvogel L, Amigorena S. Nat Rev Immunol. 2002;2:569–579. - PubMed
-
- Pan BT, Johnstone RM. Cell. 1983;33:967–977. - PubMed
-
- Fevrier B, Raposo G. Curr Opin Cell Biol. 2004;16:415–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

