FIDEL-a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut
- PMID: 20127167
- PMCID: PMC2844528
- DOI: 10.1007/s10577-009-9109-z
FIDEL-a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut
Abstract
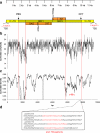
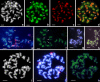
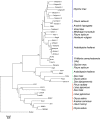
In this paper, we describe a Ty3-gypsy retrotransposon from allotetraploid peanut (Arachis hypogaea) and its putative diploid ancestors Arachis duranensis (A-genome) and Arachis ipaënsis (B-genome). The consensus sequence is 11,223 bp. The element, named FIDEL (Fairly long Inter-Dispersed Euchromatic LTR retrotransposon), is more frequent in the A- than in the B-genome, with copy numbers of about 3,000 (+/-950, A. duranensis), 820 (+/-480, A. ipaënsis), and 3,900 (+/-1,500, A. hypogaea) per haploid genome. Phylogenetic analysis of reverse transcriptase sequences showed distinct evolution of FIDEL in the ancestor species. Fluorescent in situ hybridization revealed disperse distribution in euchromatin and absence from centromeres, telomeric regions, and the nucleolar organizer region. Using paired sequences from bacterial artificial chromosomes, we showed that elements appear less likely to insert near conserved ancestral genes than near the fast evolving disease resistance gene homologs. Within the Ty3-gypsy elements, FIDEL is most closely related with the Athila/Calypso group of retrovirus-like retrotransposons. Putative transmembrane domains were identified, supporting the presence of a vestigial envelope gene. The results emphasize the importance of FIDEL in the evolution and divergence of different Arachis genomes and also may serve as an example of the role of retrotransposons in the evolution of legume genomes in general.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms—583 new estimates. Ann of Bot (Lond) 1997;80:169–196. doi: 10.1006/anbo.1997.0415. - DOI
-
- Bennett MD, Leitch IJ (2004) Plant DNA C-values database (release 3.0, Dec. 2004). http://www.kew.org/cvalues/homepage.html
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

