Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps
- PMID: 20127684
- PMCID: PMC2879143
- DOI: 10.1002/pmic.200900412
Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps
Abstract
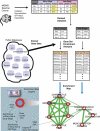
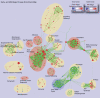
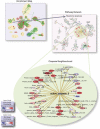
Global protein expression profiling can potentially uncover perturbations associated with common forms of heart disease. We have used shotgun MS/MS to monitor the state of biological systems in cardiac tissue correlating with disease onset, cardiac insufficiency and progression to heart failure in a time-course mouse model of dilated cardiomyopathy. However, interpreting the functional significance of the hundreds of differentially expressed proteins has been challenging. Here, we utilize improved enrichment statistical methods and an extensive collection of functionally related gene sets, gaining a more comprehensive understanding of the progressive alterations associated with functional decline in dilated cardiomyopathy. We visualize the enrichment results as an Enrichment Map, where significant gene sets are grouped based on annotation similarity. This approach vastly simplifies the interpretation of the large number of enriched gene sets found. For pathways of specific interest, such as Apoptosis and the MAPK (mitogen-activated protein kinase) cascade, we performed a more detailed analysis of the underlying signaling network, including experimental validation of expression patterns.
Figures


References
-
- Rosamond W, Flegal K, Furie K, Go A, et al. Heart Disease and Stroke Statistics 2008 Update. A Report From the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–e146. - PubMed
-
- Malcom J, Arnold O, Howlett JG, Ducharme A, et al. Canadian Cardiovascular Society Consensus Conference guidelines on heart failure – 2008 update: best practices for the transition of care of heart failure patients, and the recognition, investigation and treatment of cardiomyopathies. Can. J. Cardiol. 2008;24:21–40. - PMC - PubMed
-
- Arab S, Gramolini AO, Ping P, Kislinger T, et al. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J. Am. Coll. Cardiol. 2006;48:1733–1741. - PubMed
-
- Isserlin R, Emili A. Interpretation of large-scale quantitative shotgun proteomic profiles for biomarker discovery. Curr. Opin. Mol. Ther. 2008;10:231–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

