Genomic and small RNA sequencing of Miscanthus x giganteus shows the utility of sorghum as a reference genome sequence for Andropogoneae grasses
- PMID: 20128909
- PMCID: PMC2872872
- DOI: 10.1186/gb-2010-11-2-r12
Genomic and small RNA sequencing of Miscanthus x giganteus shows the utility of sorghum as a reference genome sequence for Andropogoneae grasses
Abstract
Background: Miscanthus x giganteus (Mxg) is a perennial grass that produces superior biomass yields in temperate environments. The essentially uncharacterized triploid genome (3n = 57, x = 19) of Mxg is likely critical for the rapid growth of this vegetatively propagated interspecific hybrid.
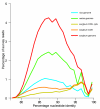
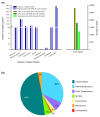
Results: A survey of the complex Mxg genome was conducted using 454 pyrosequencing of genomic DNA and Illumina sequencing-by-synthesis of small RNA. We found that the coding fraction of the Mxg genome has a high level of sequence identity to that of other grasses. Highly repetitive sequences representing the great majority of the Mxg genome were predicted using non-cognate assembly for de novo repeat detection. Twelve abundant families of repeat were observed, with those related to either transposons or centromeric repeats likely to comprise over 95% of the genome. Comparisons of abundant repeat sequences to a small RNA survey of three Mxg organs (leaf, rhizome, inflorescence) revealed that the majority of observed 24-nucleotide small RNAs are derived from these repetitive sequences. We show that high-copy-number repeats match more of the small RNA, even when the amount of the repeat sequence in the genome is accounted for.
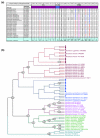
Conclusions: We show that major repeats are present within the triploid Mxg genome and are actively producing small RNAs. We also confirm the hypothesized origins of Mxg, and suggest that while the repeat content of Mxg differs from sorghum, the sorghum genome is likely to be of utility in the assembly of a gene-space sequence of Mxg.
Figures



References
-
- Brown RH. A difference in N use efficiency in C3 and C4 plants and its implications in adaptation and evolution. Crop Sci. 1978;18:93–98.
-
- Beadle CL, Long SP. Photosynthesis - is it limiting to biomass production. Biomass. 1985;8:119–168. doi: 10.1016/0144-4565(85)90022-8. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

