The hearing gene Prestin unites echolocating bats and whales
- PMID: 20129037
- PMCID: PMC11646320
- DOI: 10.1016/j.cub.2009.11.042
The hearing gene Prestin unites echolocating bats and whales
Abstract
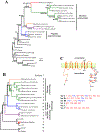
Echolocation is a sensory mechanism for locating, ranging and identifying objects which involves the emission of calls into the environment and listening to the echoes returning from objects [1]. Only microbats and toothed whales have acquired sophisticated echolocation, indispensable for their orientation and foraging [1]. Although the bat and whale biosonars originated independently and differ substantially in many aspects [2], we here report the surprising finding that the bottlenose dolphin, a toothed whale, is clustered with microbats in the gene tree constructed using protein sequences encoded by the hearing gene Prestin.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Jones G. (2005). Echolocation. Curr Biol 15, R484–488. - PubMed
-
- Au WWL (2004). A comparison of the sonar capabilities of bats and dolphins. In Echolocation in Bats and Dolphins, Thomas JA, Moss CF and Vater M, eds. (Chicago: The University of Chicago Press; ), pp. xiii–xxvii.
-
- Nei M, and Kumar S (2000). Molecular Evolution and Phylogenetics, (New York: Oxford University Press; ).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

