A human PMS2 homologue from Aquifex aeolicus stimulates an ATP-dependent DNA helicase
- PMID: 20129926
- PMCID: PMC2856984
- DOI: 10.1074/jbc.M109.050955
A human PMS2 homologue from Aquifex aeolicus stimulates an ATP-dependent DNA helicase
Abstract
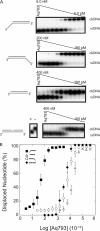
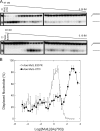
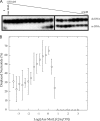
Mismatch repair in Escherichia coli involves a number of proteins including MutL and UvrD. Eukaryotes also possess MutL homologues; however, no UvrD helicase homologues have been identified. The hyperthermophilic bacterium Aquifex aeolicus has a MutL protein (Aae MutL) that possesses a latent endonuclease activity similar to eukaryotic, but different from E. coli, MutL proteins. By sequence homology Aq793 is a member of the PcrA/UvrD/Rep helicase subfamily. We expressed Aae MutL and the putative A. aeolicus DNA helicase (Aq793) proteins in E. coli. Using synthetic oligonucleotide substrates, we observed that lower concentrations of Aq793 were required to unwind double-stranded DNA that had a 3'-poly(dT) overhang as compared with double-stranded DNA with a 5'-poly(dT) or lacking a poly(dT) tail. This unwinding activity was stimulated by adding Aae MutL with maximal stimulation observed at an approximately 1.5:1 (MutL:Aq793) stoichiometric ratio. The enhancement of Aq793 helicase activity did not require the Aae MutL protein to retain endonuclease activity. Furthermore, the C-terminal 123 amino acid residues of Aae MutL were sufficient to stimulate Aq793 helicase activity, albeit at a significantly reduced efficacy. To the best of our knowledge this is the first time a human PMS2 homologue has been demonstrated to stimulate a PcrA/UvrD/Rep subfamily helicase, and this finding may further our understanding of the evolution of the mismatch repair pathway.
Figures
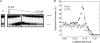
Similar articles
-
Adenosine triphosphate stimulates Aquifex aeolicus MutL endonuclease activity.PLoS One. 2009 Sep 24;4(9):e7175. doi: 10.1371/journal.pone.0007175. PLoS One. 2009. PMID: 19777055 Free PMC article.
-
MutL-catalyzed ATP hydrolysis is required at a post-UvrD loading step in methyl-directed mismatch repair.J Biol Chem. 2006 Jul 21;281(29):19949-59. doi: 10.1074/jbc.M601604200. Epub 2006 May 10. J Biol Chem. 2006. PMID: 16690604
-
Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification.J Biol Chem. 2005 Aug 12;280(32):28952-8. doi: 10.1074/jbc.M503096200. Epub 2005 Jun 13. J Biol Chem. 2005. PMID: 15955821 Free PMC article.
-
Structure and function of mismatch repair proteins.Mutat Res. 2000 Aug 30;460(3-4):245-56. doi: 10.1016/s0921-8777(00)00030-6. Mutat Res. 2000. PMID: 10946232 Review.
-
[Homologs of MutS and MutL during mammalian meiosis].Med Sci (Paris). 2003 Jan;19(1):85-91. doi: 10.1051/medsci/200319185. Med Sci (Paris). 2003. PMID: 12836196 Review. French.
Cited by
-
Plasmodium falciparum UvrD helicase translocates in 3' to 5' direction, colocalizes with MLH and modulates its activity through physical interaction.PLoS One. 2012;7(11):e49385. doi: 10.1371/journal.pone.0049385. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185322 Free PMC article.
-
Improving Performance of a SARS-CoV-2 RT-LAMP Assay for Use With a Portable Isothermal Fluorimeter: Towards a Point-of-Care Molecular Testing Strategy.J Biomol Tech. 2021 Sep;32(3):180-185. doi: 10.7171/jbt.21-3203-013. J Biomol Tech. 2021. PMID: 35027875 Free PMC article.
-
Inactivation of the DNA repair genes mutS, mutL or the anti-recombination gene mutS2 leads to activation of vitamin B1 biosynthesis genes.PLoS One. 2011 Apr 28;6(4):e19053. doi: 10.1371/journal.pone.0019053. PLoS One. 2011. PMID: 21552516 Free PMC article.
-
Novel Anti-CRISPR-Assisted CRISPR Biosensor for Exclusive Detection of Single-Stranded DNA (ssDNA).ACS Sens. 2024 Mar 22;9(3):1162-1167. doi: 10.1021/acssensors.4c00201. Epub 2024 Mar 5. ACS Sens. 2024. PMID: 38442486 Free PMC article.
-
Strand discrimination in DNA mismatch repair.DNA Repair (Amst). 2021 Sep;105:103161. doi: 10.1016/j.dnarep.2021.103161. Epub 2021 Jun 19. DNA Repair (Amst). 2021. PMID: 34171627 Free PMC article. Review.
References
-
- Modrich P., Lahue R. (1996) Annu. Rev. Biochem. 65, 101–133 - PubMed
-
- Schofield M. J., Hsieh P. (2003) Annu. Rev. Microbiol. 57, 579–608 - PubMed
-
- Harfe B. D., Jinks-Robertson S. (2000) Annu. Rev. Genet. 34, 359–399 - PubMed
-
- Kolodner R. (1996) Genes Dev. 10, 1433–1442 - PubMed
-
- Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

