Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1
- PMID: 20129943
- PMCID: PMC2879514
- DOI: 10.1093/nar/gkq033
Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1
Abstract
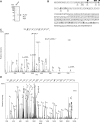
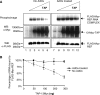
The REF/ALY mRNA export adaptor binds TAP/NXF1 via an arginine-rich region, which overlaps with its RNA-binding domain. When TAP binds a REF:RNA complex, it triggers transfer of the RNA from REF to TAP. Here, we have examined the effects of arginine methylation on the activities of the REF protein in mRNA export. We have mapped the arginine methylation sites of REF using mass spectrometry and find that several arginines within the TAP and RNA binding domains are methylated in vivo. However, arginine methylation has no effect on the REF:TAP interaction. Instead, arginine methylation reduces the RNA-binding activity of REF in vitro and in vivo. The reduced RNA-binding activity of REF in its methylated state is essential for efficient displacement of RNA from REF by TAP in vivo. Therefore, arginine methylation fine-tunes the RNA-binding activity of REF such that the RNA-protein interaction can be readily disrupted by export factors further down the pathway.
Figures



References
-
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell. Biol. 2007;8:761–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

