Dynamics of influenza virus infection and pathology
- PMID: 20130053
- PMCID: PMC2849502
- DOI: 10.1128/JVI.02078-09
Dynamics of influenza virus infection and pathology
Abstract
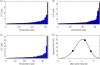
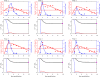
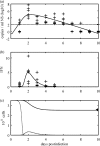
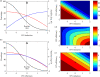
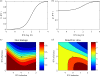
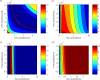
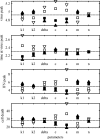
A key question in pandemic influenza is the relative roles of innate immunity and target cell depletion in limiting primary infection and modulating pathology. Here, we model these interactions using detailed data from equine influenza virus infection, combining viral and immune (type I interferon) kinetics with estimates of cell depletion. The resulting dynamics indicate a powerful role for innate immunity in controlling the rapid peak in virus shedding. As a corollary, cells are much less depleted than suggested by a model of human influenza based only on virus-shedding data. We then explore how differences in the influence of viral proteins on interferon kinetics can account for the observed spectrum of virus shedding, immune response, and influenza pathology. In particular, induction of high levels of interferon ("cytokine storms"), coupled with evasion of its effects, could lead to severe pathology, as hypothesized for some fatal cases of influenza.
Figures


References
-
- Ada, G. L., and P. D. Jones. 1986. The immune response to influenza infection. Curr. Top. Microbiol. Immunol. 128:1-54. - PubMed
-
- Beauchemin, C., J. Samuel, and J. Tuszynski. 2005. A simple cellular automaton model for influenza A viral infections. J. Theor. Biol. 232:223-234. - PubMed
-
- Bocharov, G. A., and A. A. Romanyukha. 1994. Mathematical model of antiviral immune response. III. Influenza A virus infection. J. Theor. Biol. 167:323-360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

