Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control
- PMID: 20130083
- PMCID: PMC2847520
- DOI: 10.1091/mbc.e09-10-0910
Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control
Abstract
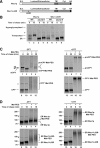
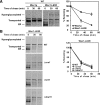
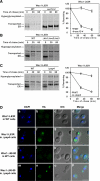
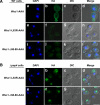
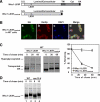
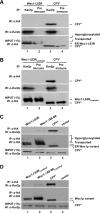
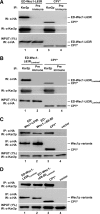
In the endoplasmic reticulum (ER), most newly synthesized proteins are retained by quality control mechanisms until folded. Misfolded molecules are sorted to ER-associated degradation (ERAD) pathways for disposal. Reports of mutant proteins degraded in the vacuole/lysosome suggested an independent Golgi-based mechanism also at work. Although little is understood of the post-ER pathway, the growing number of variants using it suggests a major role in quality control. Why seemingly redundant mechanisms in sequential compartments are needed is unclear. To understand their physiological relationship, the identification of endogenous pathway-specific substrates is a prerequisite. With ERAD substrates already well characterized, the discovery of Wsc1p as an obligate substrate of Golgi quality control enabled detailed cross-pathway analyses for the first time. By analyzing a panel of engineered substrates, the data show that the surveillance mode is determined by each polypeptide's intrinsic design. Although most secretory pathway proteins can display ERAD determinants when misfolded, the lack thereof shields Wsc1p from inspection by ER surveillance. Additionally, a powerful ER export signal mediates transport whether the luminal domain is folded or not. By evading ERAD through these passive and active mechanisms, Wsc1p is fully dependent on the post-ER system for its quality control.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

