The role of the LAT-PLC-gamma1 interaction in T regulatory cell function
- PMID: 20130215
- PMCID: PMC3646363
- DOI: 10.4049/jimmunol.0902876
The role of the LAT-PLC-gamma1 interaction in T regulatory cell function
Abstract
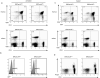
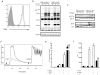
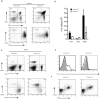
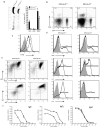
The interaction between the linker for activation of T cells (LAT) with PLC-gamma1 is important for TCR-mediated Ca(2+) signaling and MAPK activation. Knock-in mice harboring a mutation at the PLC-gamma1 binding site (Y136) of LAT develop a severe lymphoproliferative syndrome. These mice have defective thymic development and selection and lack natural regulatory T cells, implicating a breakdown of both central and peripheral tolerance. To bypass this developmental defect, we developed a conditional knock-in line in which only LATY136F is expressed in mature T cells after deletion of the wild type LAT allele. Analysis of LATY136F T cells indicated that the interaction between LAT and PLC-gamma1 plays an important role in TCR-mediated signaling, proliferation, and IL-2 production. Furthermore, the deletion of LAT induced development of the lymphoproliferative syndrome in these mice. Although Foxp3(+) natural Treg cells were present in these mice after deletion, they were unable to suppress the proliferation of conventional T cells. Our data indicate that the binding of LAT to PLC-gamma1 is essential for the suppressive function of CD4(+)CD25(+) regulatory T cells.
Figures
Similar articles
-
The importance of LAT in the activation, homeostasis, and regulatory function of T cells.J Biol Chem. 2010 Nov 12;285(46):35393-405. doi: 10.1074/jbc.M110.145052. Epub 2010 Sep 13. J Biol Chem. 2010. PMID: 20837489 Free PMC article.
-
Mutation of the phospholipase C-gamma1-binding site of LAT affects both positive and negative thymocyte selection.J Exp Med. 2005 Apr 4;201(7):1125-34. doi: 10.1084/jem.20041869. Epub 2005 Mar 28. J Exp Med. 2005. PMID: 15795236 Free PMC article.
-
LAT alleviates Th2/Treg imbalance in an OVA-induced allergic asthma mouse model through LAT-PLC-γ1 interaction.Int Immunopharmacol. 2017 Mar;44:9-15. doi: 10.1016/j.intimp.2016.12.029. Epub 2017 Jan 4. Int Immunopharmacol. 2017. PMID: 28063403
-
Lymphoproliferative disorders involving T helper effector cells with defective LAT signalosomes.Semin Immunopathol. 2010 Jun;32(2):117-25. doi: 10.1007/s00281-009-0195-y. Epub 2010 Jan 28. Semin Immunopathol. 2010. PMID: 20107804 Review.
-
LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways.Sci STKE. 2000 Dec 19;2000(63):re1. doi: 10.1126/stke.2000.63.re1. Sci STKE. 2000. PMID: 11752630 Review.
Cited by
-
The importance of LAT in the activation, homeostasis, and regulatory function of T cells.J Biol Chem. 2010 Nov 12;285(46):35393-405. doi: 10.1074/jbc.M110.145052. Epub 2010 Sep 13. J Biol Chem. 2010. PMID: 20837489 Free PMC article.
-
Ssu72 is a T-cell receptor-responsive modifier that is indispensable for regulatory T cells.Cell Mol Immunol. 2021 Jun;18(6):1395-1411. doi: 10.1038/s41423-021-00671-2. Epub 2021 Apr 13. Cell Mol Immunol. 2021. PMID: 33850312 Free PMC article.
-
Interface of Phospholipase Activity, Immune Cell Function, and Atherosclerosis.Biomolecules. 2020 Oct 15;10(10):1449. doi: 10.3390/biom10101449. Biomolecules. 2020. PMID: 33076403 Free PMC article. Review.
-
Integrative biology of T cell activation.Nat Immunol. 2014 Sep;15(9):790-7. doi: 10.1038/ni.2959. Nat Immunol. 2014. PMID: 25137453 Review.
-
Protective and detrimental roles for regulatory T cells in a viral model for multiple sclerosis.Brain Pathol. 2014 Sep;24(5):436-51. doi: 10.1111/bpa.12119. Epub 2014 Feb 25. Brain Pathol. 2014. PMID: 24417588 Free PMC article.
References
-
- Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. - PubMed
-
- Weber JR, Orstavik S, Torgersen KM, Danbolt NC, Berg SF, Ryan JC, Tasken K, Imboden JB, Vaage JT. Molecular cloning of the cDNA encoding pp36, a tyrosine-phosphorylated adaptor protein selectively expressed by T cells and natural killer cells. The Journal of experimental medicine. 1998;187:1157–1161. - PMC - PubMed
-
- Zhang W, Irvin BJ, Trible RP, Abraham RT, Samelson LE. Functional analysis of LAT in TCR-mediated signaling pathways using a LAT-deficient Jurkat cell line. International immunology. 1999;11:943–950. - PubMed
-
- Zhang W, Trible RP, Zhu M, Liu SK, McGlade CJ, Samelson LE. Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell angigen receptor-mediated signaling. The Journal of biological chemistry. 2000;275:23355–23361. - PubMed
-
- Finco TS, Kadlecek T, Zhang W, Samelson LE, Weiss A. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity. 1998;9:617–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

