Critical roles for c-Myb in lymphoid priming and early B-cell development
- PMID: 20130238
- PMCID: PMC2854426
- DOI: 10.1182/blood-2009-08-239210
Critical roles for c-Myb in lymphoid priming and early B-cell development
Abstract
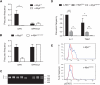
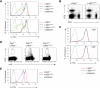
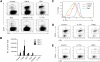
c-Myb is a transcription factor with functions in many hematopoietic lineages. c-Myb-deficient mice display reduced numbers of B cells; however, it is unknown what role c-Myb plays in B lymphopoiesis because no critical target genes have been identified in the B-cell lineage. We demonstrate that conditional deletion of c-Myb in B-cell progenitors completely abolishes B-cell development. c-Myb is required for lymphoid progenitors to respond to the cytokines interleukin-7 and thymic stromal lymphopoietin; in the absence of sufficient c-Myb activity, mice display a B lymphopenia that closely resembles that observed in interleukin-7 receptor alpha-deficient animals. Analysis of the multipotent progenitor compartment indicates that c-Myb is also required for up-regulation of multiple lymphoid-associated genes, including Il7r, and for the subsequent development of the common lymphoid progenitor population. These data show that c-Myb plays a critical role in the regulatory pathways governing lymphoid specification and early B-cell differentiation.
Figures




References
-
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1(8):661–673. - PubMed
-
- Adolfsson J, Mansson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. - PubMed
-
- Mansson R, Hultquist A, Luc S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26(4):407–419. - PubMed
-
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

