An aspartyl protease directs malaria effector proteins to the host cell
- PMID: 20130643
- PMCID: PMC2818761
- DOI: 10.1038/nature08728
An aspartyl protease directs malaria effector proteins to the host cell
Abstract
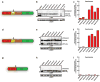
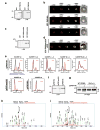
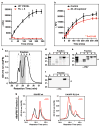
Plasmodium falciparum causes the virulent form of malaria and disease manifestations are linked to growth inside infected erythrocytes. To survive and evade host responses the parasite remodels the erythrocyte by exporting several hundred effector proteins beyond the surrounding parasitophorous vacuole membrane. A feature of exported proteins is a pentameric motif (RxLxE/Q/D) that is a substrate for an unknown protease. Here we show that the protein responsible for cleavage of this motif is plasmepsin V (PMV), an aspartic acid protease located in the endoplasmic reticulum. PMV cleavage reveals the export signal (xE/Q/D) at the amino terminus of cargo proteins. Expression of an identical mature protein with xQ at the N terminus generated by signal peptidase was not exported, demonstrating that PMV activity is essential and linked with other key export events. Identification of the protease responsible for export into erythrocytes provides a novel target for therapeutic intervention against this devastating disease.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

