Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
- PMID: 20130653
- PMCID: PMC2818718
- DOI: 10.1038/nature08722
Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Abstract
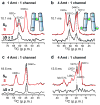
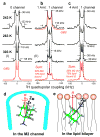
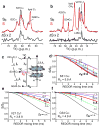
The M2 protein of influenza A virus is a membrane-spanning tetrameric proton channel targeted by the antiviral drugs amantadine and rimantadine. Resistance to these drugs has compromised their effectiveness against many influenza strains, including pandemic H1N1. A recent crystal structure of M2(22-46) showed electron densities attributed to a single amantadine in the amino-terminal half of the pore, indicating a physical occlusion mechanism for inhibition. However, a solution NMR structure of M2(18-60) showed four rimantadines bound to the carboxy-terminal lipid-facing surface of the helices, suggesting an allosteric mechanism. Here we show by solid-state NMR spectroscopy that two amantadine-binding sites exist in M2 in phospholipid bilayers. The high-affinity site, occupied by a single amantadine, is located in the N-terminal channel lumen, surrounded by residues mutated in amantadine-resistant viruses. Quantification of the protein-amantadine distances resulted in a 0.3 A-resolution structure of the high-affinity binding site. The second, low-affinity, site was observed on the C-terminal protein surface, but only when the drug reaches high concentrations in the bilayer. The orientation and dynamics of the drug are distinct in the two sites, as shown by (2)H NMR. These results indicate that amantadine physically occludes the M2 channel, thus paving the way for developing new antiviral drugs against influenza viruses. The study demonstrates the ability of solid-state NMR to elucidate small-molecule interactions with membrane proteins and determine high-resolution structures of their complexes.
Figures
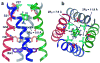
Similar articles
-
Structural basis for the function and inhibition of an influenza virus proton channel.Nature. 2008 Jan 31;451(7178):596-9. doi: 10.1038/nature06528. Nature. 2008. PMID: 18235504 Free PMC article.
-
Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.J Mol Biol. 2009 Jan 30;385(4):1127-41. doi: 10.1016/j.jmb.2008.11.022. Epub 2008 Nov 24. J Mol Biol. 2009. PMID: 19061899 Free PMC article.
-
Binding hot spots and amantadine orientation in the influenza a virus M2 proton channel.Biophys J. 2009 Nov 18;97(10):2846-53. doi: 10.1016/j.bpj.2009.09.004. Biophys J. 2009. PMID: 19917240 Free PMC article.
-
Viral M2 ion channel protein: a promising target for anti-influenza drug discovery.Mini Rev Med Chem. 2014;14(10):819-30. Mini Rev Med Chem. 2014. PMID: 25342196 Review.
-
Structural and energetic analysis of drug inhibition of the influenza A M2 proton channel.Trends Pharmacol Sci. 2013 Oct;34(10):571-80. doi: 10.1016/j.tips.2013.08.003. Epub 2013 Sep 6. Trends Pharmacol Sci. 2013. PMID: 24011996 Review.
Cited by
-
Adamantyl Isothiocyanates as Mutant p53 Rescuing Agents and Their Structure-Activity Relationships.J Med Chem. 2021 May 27;64(10):6621-6633. doi: 10.1021/acs.jmedchem.0c01971. Epub 2021 May 7. J Med Chem. 2021. PMID: 33961435 Free PMC article.
-
Residue-specific membrane location of peptides and proteins using specifically and extensively deuterated lipids and ¹³C-²H rotational-echo double-resonance solid-state NMR.J Biomol NMR. 2013 Jan;55(1):11-7. doi: 10.1007/s10858-012-9692-8. Epub 2012 Dec 8. J Biomol NMR. 2013. PMID: 23225071 Free PMC article.
-
Membrane protein structure and dynamics from NMR spectroscopy.Annu Rev Phys Chem. 2012;63:1-24. doi: 10.1146/annurev-physchem-032511-143731. Epub 2011 Nov 28. Annu Rev Phys Chem. 2012. PMID: 22136620 Free PMC article. Review.
-
Structure and Drug Binding of the SARS-CoV-2 Envelope Protein in Phospholipid Bilayers.Res Sq [Preprint]. 2020 Sep 24:rs.3.rs-77124. doi: 10.21203/rs.3.rs-77124/v1. Res Sq. 2020. Update in: Nat Struct Mol Biol. 2020 Dec;27(12):1202-1208. doi: 10.1038/s41594-020-00536-8. PMID: 32995764 Free PMC article. Updated. Preprint.
-
3D DUMAS: simultaneous acquisition of three-dimensional magic angle spinning solid-state NMR experiments of proteins.J Magn Reson. 2012 Jul;220:79-84. doi: 10.1016/j.jmr.2012.04.006. Epub 2012 Apr 26. J Magn Reson. 2012. PMID: 22698806 Free PMC article.
References
-
- Pinto LH, Lamb RA. The M2 Proton Channels of Influenza A and B Viruses. J Biol Chem. 2006;281:8997–9000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

