Crystal Structural and Functional Analysis of the Putative Dipeptidase from Pyrococcus horikoshii OT3
- PMID: 20130794
- PMCID: PMC2814137
- DOI: 10.1155/2009/434038
Crystal Structural and Functional Analysis of the Putative Dipeptidase from Pyrococcus horikoshii OT3
Abstract
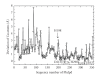
The crystal structure of a putative dipeptidase (Phdpd) from Pyrococcus horikoshii OT3 was solved using X-ray data at 2.4 A resolution. The protein is folded into two distinct entities. The N-terminal domain consists of the general topology of the alpha/beta fold, and the C-terminal domain consists of five long mixed strands, four helices, and two 3(10) helices. The structure of Phdpd is quite similar to reported structures of prolidases from P. furiosus (Zn-Pfprol) and P. horikoshii (Zn-Phdpd), where Zn ions are observed in the active site resulting in an inactive form. However, Phdpd did not contain metals in the crystal structure and showed prolidase activity in the absence of additional Co ions, whereas the specific activities increased by 5 times in the presence of a sufficient concentration (1.2 mM) of Co ions. The substrate specificities (X-Pro) of Phdpd were broad compared with those of Zn-Phdpd in the presence of Co ions, whose relative activities are 10% or less for substrates other than Met-Pro, which is the most favorable substrate. The binding constants of Zn-Phdpd with three metals (Zn, Co, and Mn) were higher than those of Phdpd and that with Zn was higher by greater than 2 orders, which were determined by DSC experiments. From the structural comparison of both forms and the above experimental results, it could be elucidated why the protein with Zn(2+) ions is inactive.
Figures



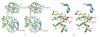

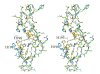
Similar articles
-
Structure of the prolidase from Pyrococcus furiosus.Biochemistry. 2004 Mar 16;43(10):2771-83. doi: 10.1021/bi0356451. Biochemistry. 2004. PMID: 15005612
-
Crystal structure and structural stability of acylphosphatase from hyperthermophilic archaeon Pyrococcus horikoshii OT3.Proteins. 2005 Oct 1;61(1):196-205. doi: 10.1002/prot.20535. Proteins. 2005. PMID: 16080154
-
Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme.J Mol Biol. 2006 Mar 24;357(2):583-91. doi: 10.1016/j.jmb.2005.12.086. Epub 2006 Jan 11. J Mol Biol. 2006. PMID: 16430919
-
Characterization of native and recombinant forms of an unusual cobalt-dependent proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus.J Bacteriol. 1998 Sep;180(18):4781-9. doi: 10.1128/JB.180.18.4781-4789.1998. J Bacteriol. 1998. PMID: 9733678 Free PMC article.
-
A three-dimensional model of RNase P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3.Biochem Biophys Res Commun. 2017 Nov 18;493(2):1063-1068. doi: 10.1016/j.bbrc.2017.09.085. Epub 2017 Sep 19. Biochem Biophys Res Commun. 2017. PMID: 28935369
Cited by
-
High-level expression and molecular characterization of a recombinant prolidase from Escherichia coli NovaBlue.PeerJ. 2018 Oct 31;6:e5863. doi: 10.7717/peerj.5863. eCollection 2018. PeerJ. 2018. PMID: 30402354 Free PMC article.
-
Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Nov 1;68(Pt 11):1275-8. doi: 10.1107/S174430911203761X. Epub 2012 Oct 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 23143231 Free PMC article.
-
Crystallization and preliminary X-ray diffraction analysis of Xaa-Pro dipeptidase from Xanthomonas campestris.Acta Crystallogr F Struct Biol Commun. 2014 Sep;70(Pt 9):1268-71. doi: 10.1107/S2053230X14017324. Epub 2014 Aug 27. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25195907 Free PMC article.
-
Effect of heavy atoms on the thermal stability of α-amylase from Aspergillus oryzae.PLoS One. 2013;8(2):e57432. doi: 10.1371/journal.pone.0057432. Epub 2013 Feb 25. PLoS One. 2013. PMID: 23451229 Free PMC article.
-
Improving the catalytic activity of hyperthermophilic Pyrococcus horikoshii prolidase for detoxification of organophosphorus nerve agents over a broad range of temperatures.Archaea. 2011;2011:565127. doi: 10.1155/2011/565127. Epub 2011 Nov 28. Archaea. 2011. PMID: 22162664 Free PMC article.
References
-
- Booth M, Jennings PV, Fhaolain IN, O'Cuinn G. Prolidase activity of Lactobacillus lactis subsp. cremoris AM2: partial purification and characterization. Journal of Dairy Research. 1990;57:245–254.
-
- Browne P, O'Cuinn G. The purification and characterization of a proline dipeptidase from guinea pig brain. The Journal of Biological Chemistry. 1983;258(10):6147–6154. - PubMed
-
- Endo F, Tanoue A, Nakai H, et al. Primary structure and gene localization of human prolidase. The Journal of Biological Chemistry. 1989;264(8):4476–4481. - PubMed
-
- Sjöström H, Norén O, Josefsson L. Purification and specificity of pig intestinal prolidase. Biochimica et Biophysica Acta. 1973;327(2):457–470. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

