Ordered transcriptional factor recruitment and epigenetic regulation of tnf-alpha in necrotizing acute pancreatitis
- PMID: 20130956
- PMCID: PMC11115704
- DOI: 10.1007/s00018-010-0272-3
Ordered transcriptional factor recruitment and epigenetic regulation of tnf-alpha in necrotizing acute pancreatitis
Abstract
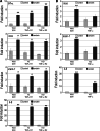
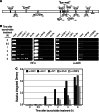
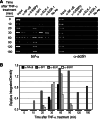
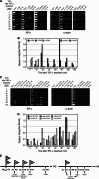
Tauhe expression of the critical initiator cytokine TNF-alpha was strongly upregulated in vivo in acute necrotic pancreatitis (AP) in rodents and in vitro in TNF-alpha activated acinar AR42J cells. Upregulation of tnf-alpha, inos, icam-1 and il-6 occurred both in TNF-alpha receptor 1 and 2 knock-out mice, but not in TNF-alpha knock-out mice, in cerulein-induced acute pancreatitis. Chromatin immunoprecipitation analysis showed that transcriptional factors (ELK-1, SP1, NF-kappaB and EGR-1) and chromatin modification complexes (HDAC1, HDAC2, GCN5, PCAF and CBP) were recruited and/or released from the promoter in a strictly ordered mechanism. Activation of tnf-alpha gene was also accompanied by an ordered increased level of histone H3K9, H3K14 and H3K18-acetylation and H3K4 methylation, as well as H4K5 acetylation. A better knowledge of the molecular mechanisms that control tnf-alpha gene regulation will provide deeper understanding of the initiation and development of the inflammatory processes occurring in acute pancreatitis triggered by TNF-alpha cytokine.
Figures


Similar articles
-
Epigenetic Regulation of Early- and Late-Response Genes in Acute Pancreatitis.J Immunol. 2016 Nov 15;197(10):4137-4150. doi: 10.4049/jimmunol.1502378. Epub 2016 Oct 19. J Immunol. 2016. PMID: 27798150
-
Synchronous recruitment of epigenetic modifiers to endotoxin synergistically activated Tnf-α gene in acute kidney injury.PLoS One. 2013 Jul 30;8(7):e70322. doi: 10.1371/journal.pone.0070322. Print 2013. PLoS One. 2013. PMID: 23936185 Free PMC article.
-
Karyopherin Alpha 2 Promotes the Inflammatory Response in Rat Pancreatic Acinar Cells Via Facilitating NF-κB Activation.Dig Dis Sci. 2016 Mar;61(3):747-57. doi: 10.1007/s10620-015-3948-6. Epub 2015 Nov 2. Dig Dis Sci. 2016. PMID: 26526450
-
Pentoxifylline prevents loss of PP2A phosphatase activity and recruitment of histone acetyltransferases to proinflammatory genes in acute pancreatitis.J Pharmacol Exp Ther. 2009 Nov;331(2):609-17. doi: 10.1124/jpet.109.157537. Epub 2009 Aug 11. J Pharmacol Exp Ther. 2009. PMID: 19671881
-
Gene deletion of MK2 inhibits TNF-alpha and IL-6 and protects against cerulein-induced pancreatitis.Am J Physiol Gastrointest Liver Physiol. 2006 Jun;290(6):G1298-306. doi: 10.1152/ajpgi.00530.2005. Epub 2006 Jan 19. Am J Physiol Gastrointest Liver Physiol. 2006. PMID: 16423921
Cited by
-
Application of nanotechnology in the diagnosis and treatment of acute pancreatitis.Nanoscale Adv. 2022 Mar 19;4(8):1949-1961. doi: 10.1039/d2na00020b. eCollection 2022 Apr 12. Nanoscale Adv. 2022. PMID: 36133408 Free PMC article. Review.
-
Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components.Infect Immun. 2010 Sep;78(9):3822-31. doi: 10.1128/IAI.00433-10. Epub 2010 Jul 6. Infect Immun. 2010. PMID: 20605984 Free PMC article.
-
Silencing of CREB Inhibits HDAC2/TLR4/NF-κB Cascade to Relieve Severe Acute Pancreatitis-Induced Myocardial Injury.Inflammation. 2021 Aug;44(4):1565-1580. doi: 10.1007/s10753-021-01441-y. Epub 2021 Mar 16. Inflammation. 2021. Retraction in: Inflammation. 2024 Aug;47(4):1546. doi: 10.1007/s10753-024-01987-7. PMID: 33725236 Retracted.
-
Epigenetic transcriptional regulation of the growth arrest-specific gene 1 (Gas1) in hepatic cell proliferation at mononucleosomal resolution.PLoS One. 2011;6(8):e23318. doi: 10.1371/journal.pone.0023318. Epub 2011 Aug 9. PLoS One. 2011. PMID: 21858068 Free PMC article.
-
CVB3 VP1 interacts with MAT1 to inhibit cell proliferation by interfering with Cdk-activating kinase complex activity in CVB3-induced acute pancreatitis.PLoS Pathog. 2021 Feb 8;17(2):e1008992. doi: 10.1371/journal.ppat.1008992. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33556114 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

