Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain
- PMID: 20131913
- PMCID: PMC2849892
- DOI: 10.1021/pr901171q
Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain
Abstract
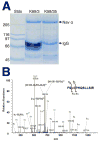
Reversible phosphorylation of ion channels underlies cellular plasticity in mammalian neurons. Voltage-gated sodium or Nav channels underlie action potential initiation and propagation, dendritic excitability, and many other aspects of neuronal excitability. Various protein kinases have been suggested to phosphorylate the primary or alpha subunit of Nav channels, affecting diverse aspects of channel function. Previous studies of Nav alpha subunit phosphorylation have led to the identification of a small set of phosphorylation sites important in mediating diverse aspects of Nav channel function. Here we use nanoflow liquid chromatography tandem mass spectrometry (nano-LC MS/MS) on Nav alpha subunits affinity-purified from rat brain with two distinct monoclonal antibodies to identify 15 phosphorylation sites on Nav1.2, 12 of which have not been previously reported. We also found 3 novel phosphorylation sites on Nav1.1. In general, commonly used phosphorylation site prediction algorithms did not accurately predict these novel in vivo phosphorylation sites. Our results demonstrate that specific Nav alpha subunits isolated from rat brain are highly phosphorylated, and suggest extensive modulation of Nav channel activity in mammalian brain. Identification of phosphorylation sites using monoclonal antibody-based immunopurification and mass spectrometry is an effective approach to define the phosphorylation status of Nav channels and other important membrane proteins in mammalian brain.
Figures




Similar articles
-
Mass spectrometry-based identification of native cardiac Nav1.5 channel α subunit phosphorylation sites.J Proteome Res. 2012 Dec 7;11(12):5994-6007. doi: 10.1021/pr300702c. Epub 2012 Nov 9. J Proteome Res. 2012. PMID: 23092124 Free PMC article.
-
Reciprocal changes in phosphorylation and methylation of mammalian brain sodium channels in response to seizures.J Biol Chem. 2014 May 30;289(22):15363-73. doi: 10.1074/jbc.M114.562785. Epub 2014 Apr 15. J Biol Chem. 2014. PMID: 24737319 Free PMC article.
-
Profiling the phospho-status of the BKCa channel alpha subunit in rat brain reveals unexpected patterns and complexity.Mol Cell Proteomics. 2008 Nov;7(11):2188-98. doi: 10.1074/mcp.M800063-MCP200. Epub 2008 Jun 23. Mol Cell Proteomics. 2008. PMID: 18573811 Free PMC article.
-
Cell-Adhesion Properties of β-Subunits in the Regulation of Cardiomyocyte Sodium Channels.Biomolecules. 2020 Jul 1;10(7):989. doi: 10.3390/biom10070989. Biomolecules. 2020. PMID: 32630316 Free PMC article. Review.
-
Molecular properties of brain sodium channels: an important target for anticonvulsant drugs.Adv Neurol. 1999;79:441-56. Adv Neurol. 1999. PMID: 10514834 Review.
Cited by
-
The Nav1.2 channel is regulated by GSK3.Biochim Biophys Acta. 2015 Apr;1850(4):832-44. doi: 10.1016/j.bbagen.2015.01.011. Epub 2015 Jan 20. Biochim Biophys Acta. 2015. PMID: 25615535 Free PMC article.
-
The fibroblast growth factor 14·voltage-gated sodium channel complex is a new target of glycogen synthase kinase 3 (GSK3).J Biol Chem. 2013 Jul 5;288(27):19370-85. doi: 10.1074/jbc.M112.445924. Epub 2013 May 2. J Biol Chem. 2013. PMID: 23640885 Free PMC article.
-
Identifying a kinase network regulating FGF14:Nav1.6 complex assembly using split-luciferase complementation.PLoS One. 2015 Feb 6;10(2):e0117246. doi: 10.1371/journal.pone.0117246. eCollection 2015. PLoS One. 2015. PMID: 25659151 Free PMC article.
-
PKCε phosphorylation of the sodium channel NaV1.8 increases channel function and produces mechanical hyperalgesia in mice.J Clin Invest. 2012 Apr;122(4):1306-15. doi: 10.1172/JCI61934. Epub 2012 Mar 19. J Clin Invest. 2012. PMID: 22426212 Free PMC article.
-
An update on transcriptional and post-translational regulation of brain voltage-gated sodium channels.Amino Acids. 2016 Mar;48(3):641-651. doi: 10.1007/s00726-015-2122-y. Epub 2015 Oct 27. Amino Acids. 2016. PMID: 26503606 Free PMC article. Review.
References
-
- Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57(4):397–409. - PubMed
-
- Losonczy A, Makara JK, Magee JC. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature. 2008;452(7186):436–41. - PubMed
-
- Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7(1):42–54. - PubMed
-
- Agnew WS. Neurochemistry. Lessons from large molecules. Nature. 1986;322(6082):770–1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

