Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases
- PMID: 20132407
- PMCID: PMC3823453
- DOI: 10.1111/j.1582-4934.2010.01018.x
Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases
Abstract
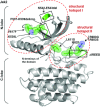
Gain-of-function mutations in the genes encoding Janus kinases have been discovered in various haematologic diseases. Jaks are composed of a FERM domain, an SH2 domain, a pseudokinase domain and a kinase domain, and a complex interplay of the Jak domains is involved in regulation of catalytic activity and association to cytokine receptors. Most activating mutations are found in the pseudokinase domain. Here we present recently discovered mutations in the context of our structural models of the respective domains. We describe two structural hotspots in the pseudokinase domain of Jak2 that seem to be associated either to myeloproliferation or to lymphoblastic leukaemia, pointing at the involvement of distinct signalling complexes in these disease settings. The different domains of Jaks are discussed as potential drug targets. We present currently available inhibitors targeting Jaks and indicate structural differences in the kinase domains of the different Jaks that may be exploited in the development of specific inhibitors. Moreover, we discuss recent chemical genetic approaches which can be applied to Jaks to better understand the role of these kinases in their biological settings and as drug targets.
Figures

Similar articles
-
Activating Janus kinase pseudokinase domain mutations in myeloproliferative and other blood cancers.Biochem Soc Trans. 2013 Aug;41(4):1048-54. doi: 10.1042/BST20130084. Biochem Soc Trans. 2013. PMID: 23863177 Review.
-
Janus kinase 2 activation mechanisms revealed by analysis of suppressing mutations.J Allergy Clin Immunol. 2019 Apr;143(4):1549-1559.e6. doi: 10.1016/j.jaci.2018.07.022. Epub 2018 Aug 6. J Allergy Clin Immunol. 2019. PMID: 30092288 Free PMC article.
-
Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2).J Biol Chem. 2015 Nov 6;290(45):27261-27270. doi: 10.1074/jbc.M115.672048. Epub 2015 Sep 10. J Biol Chem. 2015. PMID: 26359499 Free PMC article.
-
Jaks and cytokine receptors--an intimate relationship.Biochem Pharmacol. 2006 Nov 30;72(11):1538-46. doi: 10.1016/j.bcp.2006.04.013. Epub 2006 Apr 27. Biochem Pharmacol. 2006. PMID: 16750817 Review.
-
Therapeutic targeting of Janus kinases.Immunol Rev. 2008 Jun;223:132-42. doi: 10.1111/j.1600-065X.2008.00644.x. Immunol Rev. 2008. PMID: 18613833 Free PMC article. Review.
Cited by
-
Allosteric regulation and inhibition of protein kinases.Biochem Soc Trans. 2023 Feb 27;51(1):373-385. doi: 10.1042/BST20220940. Biochem Soc Trans. 2023. PMID: 36794774 Free PMC article. Review.
-
JAK2 mutants (e.g., JAK2V617F) and their importance as drug targets in myeloproliferative neoplasms.JAKSTAT. 2013 Jul 1;2(3):e25025. doi: 10.4161/jkst.25025. Epub 2013 May 14. JAKSTAT. 2013. PMID: 24069563 Free PMC article. Review.
-
The use of structural biology in Janus kinase targeted drug discovery.Curr Drug Targets. 2011 Apr;12(4):546-55. doi: 10.2174/138945011794751528. Curr Drug Targets. 2011. PMID: 21126226 Free PMC article. Review.
-
Recent advances in targeting protein kinases and pseudokinases in cancer biology.Front Cell Dev Biol. 2022 Jul 22;10:942500. doi: 10.3389/fcell.2022.942500. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35938171 Free PMC article. Review.
-
Oncogenic JAK1 and JAK2-activating mutations resistant to ATP-competitive inhibitors.Haematologica. 2011 Jun;96(6):845-53. doi: 10.3324/haematol.2010.036350. Epub 2011 Mar 10. Haematologica. 2011. PMID: 21393331 Free PMC article.
References
-
- Ortutay C, Valiaho J, Stenberg K, et al. KinMutBase: a registry of disease-causing mutations in protein kinase domains. Hum Mutat. 2005;25:435–42. - PubMed
-
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–9. - PubMed
-
- Janne PA, Johnson BE. Effect of epidermal growth factor receptor tyrosine kinase domain mutations on the outcome of patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res. 2006;12:4416s–20s. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

