Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families
- PMID: 20132533
- PMCID: PMC2832619
- DOI: 10.1186/1741-7007-8-12
Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families
Abstract
Background: Most rapidly evolving gene families are involved in immune responses and reproduction, two biological functions which have been assigned to the carcinoembryonic antigen (CEA) gene family. To gain insights into evolutionary forces shaping the CEA gene family we have analysed this gene family in 27 mammalian species including monotreme and marsupial lineages.
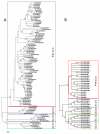
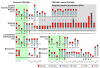
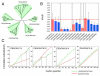
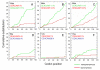
Results: Phylogenetic analysis provided convincing evidence that the primordial CEA gene family in mammals consisted of five genes, including the immune inhibitory receptor-encoding CEACAM1 (CEA-related cell adhesion molecule) ancestor. Our analysis of the substitution rates within the nucleotide sequence which codes for the ligand binding domain of CEACAM1 indicates that the selection for diversification is, perhaps, a consequence of the exploitation of CEACAM1 by a variety of viral and bacterial pathogens as their cellular receptor. Depending on the extent of the amplification of an ancestral CEACAM1, the number of CEACAM1-related genes varies considerably between mammalian species from less than five in lagomorphs to more than 100 in bats. In most analysed species, ITAM (immunoreceptor tyrosine-based activation motifs) or ITAM-like motif-containing proteins exist which contain Ig-V-like, ligand binding domains closely related to that of CEACAM1. Human CEACAM3 is one such protein which can function as a CEACAM1 decoy receptor in granulocytes by mediating the uptake and destruction of specific bacterial pathogens via its ITAM-like motif. The close relationship between CEACAM1 and its ITAM-encoding relatives appears to be maintained by gene conversion and reciprocal recombination. Surprisingly, secreted CEACAMs resembling immunomodulatory CEACAM1-related trophoblast-specific pregnancy-specific glycoproteins (PSGs) found in humans and rodents evolved only in a limited set of mammals. The appearance of PSG-like genes correlates with invasive trophoblast growth in these species.
Conclusions: These phylogenetic studies provide evidence that pathogen/host coevolution and a possible participation in fetal-maternal conflict processes led to a highly species-specific diversity of mammalian CEA gene families.
Figures




Similar articles
-
Species-specific evolution of immune receptor tyrosine based activation motif-containing CEACAM1-related immune receptors in the dog.BMC Evol Biol. 2007 Oct 18;7:196. doi: 10.1186/1471-2148-7-196. BMC Evol Biol. 2007. PMID: 17945019 Free PMC article.
-
Coevolution of paired receptors in Xenopus carcinoembryonic antigen-related cell adhesion molecule families suggests appropriation as pathogen receptors.BMC Genomics. 2016 Nov 16;17(1):928. doi: 10.1186/s12864-016-3279-9. BMC Genomics. 2016. PMID: 27852220 Free PMC article.
-
Identification of allelic variants of the bovine immune regulatory molecule CEACAM1 implies a pathogen-driven evolution.Gene. 2004 Sep 15;339:99-109. doi: 10.1016/j.gene.2004.06.023. Gene. 2004. PMID: 15363850
-
[Human carcinoembryonic antigen family proteins, structure and function].Postepy Hig Med Dosw (Online). 2012 Jul 20;66:521-33. doi: 10.5604/17322693.1004113. Postepy Hig Med Dosw (Online). 2012. PMID: 22922152 Review. Polish.
-
[CEACAM1--a less well-known member of the family of carcinoembryonic antigens].Cas Lek Cesk. 2003;142(5):259-63. Cas Lek Cesk. 2003. PMID: 12920788 Review. Czech.
Cited by
-
Mechanisms of host adaptation by bacterial pathogens.FEMS Microbiol Rev. 2024 Jun 20;48(4):fuae019. doi: 10.1093/femsre/fuae019. FEMS Microbiol Rev. 2024. PMID: 39003250 Free PMC article. Review.
-
Development and characterization of NILK-2301, a novel CEACAM5xCD3 κλ bispecific antibody for immunotherapy of CEACAM5-expressing cancers.J Hematol Oncol. 2023 Dec 12;16(1):117. doi: 10.1186/s13045-023-01516-3. J Hematol Oncol. 2023. PMID: 38087365 Free PMC article.
-
Alternative splicing after gene duplication drives CEACAM1-paralog diversification in the horse.BMC Evol Biol. 2018 Mar 15;18(1):32. doi: 10.1186/s12862-018-1145-x. BMC Evol Biol. 2018. PMID: 29544443 Free PMC article.
-
The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources.Dev Biol. 2016 Jan 1;409(1):251-260. doi: 10.1016/j.ydbio.2015.10.015. Epub 2015 Oct 23. Dev Biol. 2016. PMID: 26476147 Free PMC article.
-
The granulocyte orphan receptor CEACAM4 is able to trigger phagocytosis of bacteria.J Leukoc Biol. 2015 Mar;97(3):521-31. doi: 10.1189/jlb.2AB0813-449RR. Epub 2015 Jan 7. J Leukoc Biol. 2015. PMID: 25567962 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

