Caspase 8 and maspin are downregulated in breast cancer cells due to CpG site promoter methylation
- PMID: 20132554
- PMCID: PMC2824712
- DOI: 10.1186/1471-2407-10-32
Caspase 8 and maspin are downregulated in breast cancer cells due to CpG site promoter methylation
Abstract
Background: Epigenetic changes associated with promoter DNA methylation results in silencing of several tumor suppressor genes that lead to increased risk for tumor formation and for progression of the cancer.
Methods: Methylation specific PCR (MSP) and bisulfite sequencing were used for determination of proapoptotic gene Caspase 8 (CASP8) and the tumor suppressor gene maspin promoter methylation in four breast cancer and two non-tumorigenic breast cell lines. Involvement of histone H3 methylation in those cell lines were examined by CHIP assay.
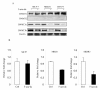
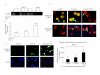
Results: The CpG sites in the promoter region of CASP8 and maspin were methylated in all four breast cancer cell lines but not in two non-tumorigenic breast cell lines. Demethylation agent 5-aza-2'-deoxycytidine (5-aza-dc) selectively inhibits DNA methyltransferases, DNMT3a and DNMT3b, and restored CASP8 and maspin gene expression in breast cancer cells. 5-aza-dc also reduced histone H3k9me2 occupancy on CASP8 promoter in SKBR3cells, but not in MCF-7 cells. Combination of histone deacetylase inhibitor Trichostatin A (TSA) and 5-aza-dc significant decrease in nuclear expression of Di-methyl histone H3-Lys27 and slight increase in acetyl histone H3-Lys9 in MCF-7 cells. CASP8 mRNA and protein level in MCF-7 cells were increased by the 5-aza-dc in combination with TSA. Data from our study also demonstrated that treatment with 5-FU caused a significant increase in unmethylated CASP8 and in CASP8 mRNA in all 3 cancer lines.
Conclusions: CASP8 and maspin expression were reduced in breast cancer cells due to promoter methylation. Selective application of demethylating agents could offer novel therapeutic opportunities in breast cancer.
Figures

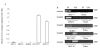


Similar articles
-
Epigenetic silencing of maspin expression occurs early in the conversion of keratocytes to fibroblasts.Exp Eye Res. 2008 Apr;86(4):586-600. doi: 10.1016/j.exer.2008.01.003. Epub 2008 Jan 12. Exp Eye Res. 2008. PMID: 18291368 Free PMC article.
-
DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac.J Exp Clin Cancer Res. 2016 Jul 26;35(1):120. doi: 10.1186/s13046-016-0396-x. J Exp Clin Cancer Res. 2016. PMID: 27461117 Free PMC article.
-
Demethylation by 5-aza-2'-deoxycytidine in colorectal cancer cells targets genomic DNA whilst promoter CpG island methylation persists.BMC Cancer. 2010 Jul 12;10:366. doi: 10.1186/1471-2407-10-366. BMC Cancer. 2010. PMID: 20618997 Free PMC article.
-
Re-expression of methylation-induced tumor suppressor gene silencing is associated with the state of histone modification in gastric cancer cell lines.World J Gastroenterol. 2007 Dec 14;13(46):6166-71. doi: 10.3748/wjg.v13.i46.6166. World J Gastroenterol. 2007. PMID: 18069755 Free PMC article.
-
Maspin: the new frontier.Clin Cancer Res. 2006 Dec 15;12(24):7279-83. doi: 10.1158/1078-0432.CCR-06-1589. Clin Cancer Res. 2006. PMID: 17189399 Free PMC article. Review.
Cited by
-
Epigenetic silencing of apoptosis-inducing gene expression can be efficiently overcome by combined SAHA and TRAIL treatment in uterine sarcoma cells.PLoS One. 2014 Mar 11;9(3):e91558. doi: 10.1371/journal.pone.0091558. eCollection 2014. PLoS One. 2014. PMID: 24618889 Free PMC article.
-
DNA methylation in thyroid tumorigenesis.Cancers (Basel). 2011 Jun 1;3(2):1732-43. doi: 10.3390/cancers3021732. Cancers (Basel). 2011. PMID: 21738852 Free PMC article.
-
Multi-omic Dissection of Oncogenically Active Epiproteomes Identifies Drivers of Proliferative and Invasive Breast Tumors.iScience. 2019 Jul 26;17:359-378. doi: 10.1016/j.isci.2019.07.001. Epub 2019 Jul 4. iScience. 2019. PMID: 31336272 Free PMC article.
-
Polympact: exploring functional relations among common human genetic variants.Nucleic Acids Res. 2022 Feb 22;50(3):1335-1350. doi: 10.1093/nar/gkac024. Nucleic Acids Res. 2022. PMID: 35061909 Free PMC article.
-
Nuclear location of tumor suppressor protein maspin inhibits proliferation of breast cancer cells without affecting proliferation of normal epithelial cells.BMC Cancer. 2014 Feb 28;14:142. doi: 10.1186/1471-2407-14-142. BMC Cancer. 2014. PMID: 24581141 Free PMC article.
References
-
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–480. - PubMed
-
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. full_text. - PubMed
-
- Miyakura Y, Sugano K, Konishi F, Ichikawa A, Maekawa M, Shitoh K, Shitoh K, Igarashi S, Kotake K, Koyama Y, Nagai H. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300–1309. doi: 10.1053/gast.2001.29616. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

