Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters
- PMID: 20133331
- PMCID: PMC2847750
- DOI: 10.1101/gr.103226.109
Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters
Abstract
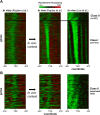
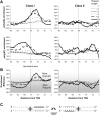
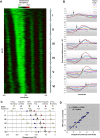
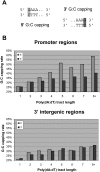
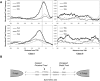
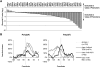
Eukaryotic transcriptional regulation is mediated by the organization of nucleosomes in promoter regions. Most Saccharomyces cerevisiae promoters have a highly stereotyped chromatin organization, where nucleosome-free regions (NFR) are flanked by well-ordered nucleosomes. We have found that yeast promoters fall into two classes differing in NFR sharpness, and that this distinction follows a known transcriptional dichotomy in yeast genes. A class of yeast promoters having well-defined NFRs are characterized by positioned patterns of poly(dA:dT) tracts with several novel features. First, poly(dA:dT) tracts are localized in a strand-dependent manner, with poly(dA) tracts lying proximal to transcriptional start sites and poly(dT) tracts lying distal, and collectively define a symmetry axis that is coincident with NFR centers. Second, poly(dA:dT) tracts are preferentially "capped" by G:C residues on the terminus proximal to the symmetry axis. Both signature features co-vary with fine positional variations between NFRs, establishing a closely knit relationship between poly(dA:dT) tracts, their capping patterns, and the central coordinates of NFRs. We found that these features are unique to promoters with well-defined NFRs, and that these promoters display significant difference between in vitro and in vivo nucleosome occupancy patterns. These observations are consistent with a model in which localized and G:C-capped poly(dA:dT) tracts initiate or facilitate the formation of NFRs at their center, possibly with chromatin remodeling and transcriptional machines involved.
Figures


Similar articles
-
The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo.Int J Mol Sci. 2021 Jul 30;22(15):8233. doi: 10.3390/ijms22158233. Int J Mol Sci. 2021. PMID: 34360997 Free PMC article. Review.
-
Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo.Nucleic Acids Res. 2000 Nov 1;28(21):4083-9. doi: 10.1093/nar/28.21.4083. Nucleic Acids Res. 2000. PMID: 11058103 Free PMC article.
-
Poly(dA:dT) Tracts Differentially Modulate Nucleosome Remodeling Activity of RSC and ISW1a Complexes, Exerting Tract Orientation-Dependent and -Independent Effects.Int J Mol Sci. 2023 Oct 17;24(20):15245. doi: 10.3390/ijms242015245. Int J Mol Sci. 2023. PMID: 37894925 Free PMC article.
-
Poly(dA).poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter.Nucleic Acids Res. 1990 Jun 25;18(12):3495-502. doi: 10.1093/nar/18.12.3495. Nucleic Acids Res. 1990. PMID: 2194162 Free PMC article.
-
Poly(dA:dT) tracts: major determinants of nucleosome organization.Curr Opin Struct Biol. 2009 Feb;19(1):65-71. doi: 10.1016/j.sbi.2009.01.004. Epub 2009 Feb 7. Curr Opin Struct Biol. 2009. PMID: 19208466 Free PMC article. Review.
Cited by
-
The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo.Int J Mol Sci. 2021 Jul 30;22(15):8233. doi: 10.3390/ijms22158233. Int J Mol Sci. 2021. PMID: 34360997 Free PMC article. Review.
-
A conserved GA element in TATA-less RNA polymerase II promoters.PLoS One. 2011;6(11):e27595. doi: 10.1371/journal.pone.0027595. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22110682 Free PMC article.
-
GC content strongly influences the role of poly(dA) in the intrinsic nucleosome positioning in Saccharomyces cerevisiae.Yeast. 2022 Apr;39(4):262-271. doi: 10.1002/yea.3701. Epub 2022 Mar 29. Yeast. 2022. PMID: 35348238 Free PMC article.
-
Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae.Genome Biol. 2020 Jun 2;21(1):132. doi: 10.1186/s13059-020-02040-0. Genome Biol. 2020. PMID: 32487207 Free PMC article.
-
Genomic Nucleosome Organization Reconstituted with Pure Proteins.Cell. 2016 Oct 20;167(3):709-721.e12. doi: 10.1016/j.cell.2016.09.045. Cell. 2016. PMID: 27768892 Free PMC article.
References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA·dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–624. - PubMed
-
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
