Regulation of alternative splicing by histone modifications
- PMID: 20133523
- PMCID: PMC2913848
- DOI: 10.1126/science.1184208
Regulation of alternative splicing by histone modifications
Abstract
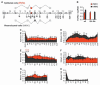
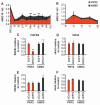
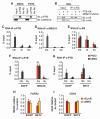
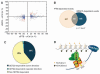
Alternative splicing of pre-mRNA is a prominent mechanism to generate protein diversity, yet its regulation is poorly understood. We demonstrated a direct role for histone modifications in alternative splicing. We found distinctive histone modification signatures that correlate with the splicing outcome in a set of human genes, and modulation of histone modifications causes splice site switching. Histone marks affect splicing outcome by influencing the recruitment of splicing regulators via a chromatin-binding protein. These results outline an adaptor system for the reading of histone marks by the pre-mRNA splicing machinery.
Figures

Comment in
-
Chromatin: the final frontier in splicing regulation?Dev Cell. 2010 Mar 16;18(3):336-8. doi: 10.1016/j.devcel.2010.03.002. Dev Cell. 2010. PMID: 20230741 Free PMC article.
-
Journal club. A human geneticist explores the ways that genes are regulated.Nature. 2010 Jul 1;466(7302):11. doi: 10.1038/466011e. Nature. 2010. PMID: 20595974 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

