Spontaneous assembly and active disassembly balance adherens junction homeostasis
- PMID: 20133579
- PMCID: PMC2840517
- DOI: 10.1073/pnas.0911027107
Spontaneous assembly and active disassembly balance adherens junction homeostasis
Abstract
The homeostasis of adherens junctions was studied using E-cadherin and its two mutants tagged by the photoconvertible protein Dendra2 in epithelial A-431 cells and in CHO cells lacking endogenous cadherin. The first mutant contained point mutations of two elements, Lys738 and the dileucine motif that suppressed cadherin endocytosis. The second mutant contained, in addition, an extensive truncation that uncoupled the mutant from beta-catenin and p120. Surprisingly, the intact cadherin and its truncated mutant were recruited into the junctions with identical kinetics. The full-size cadherin was actively removed from the junctions by a process that was unaffected by the inactivation of its endocytic elements. The cadherin's apparent half-residence time in the junction was about 2 min. Cadherin clusters made of the truncated mutant exhibited much slower but ATP-independent junctional turnover. Taken together, our experiments showed that adherens junction homeostasis consists of three distinctive steps: cadherin spontaneous recruitment, its lateral catenin-dependent association, and its active release from the resulting clusters. The latter process, whose mechanism is not clear, may play an important role in various kinds of normal and abnormal morphogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
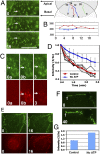

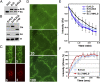
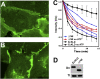
References
-
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. - PubMed
-
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. - PubMed
-
- Troyanovsky S. Cadherin dimers in cell-cell adhesion. Eur J Cell Biol. 2005;84:225–233. - PubMed
-
- Provost E, Rimm DL. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr Opin Cell Biol. 1999;11:567–572. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

