Translation efficiency is determined by both codon bias and folding energy
- PMID: 20133581
- PMCID: PMC2840511
- DOI: 10.1073/pnas.0909910107
Translation efficiency is determined by both codon bias and folding energy
Abstract
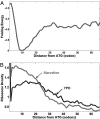
Synonymous mutations do not alter the protein produced yet can have a significant effect on protein levels. The mechanisms by which this effect is achieved are controversial; although some previous studies have suggested that codon bias is the most important determinant of translation efficiency, a recent study suggested that mRNA folding at the beginning of genes is the dominant factor via its effect on translation initiation. Using the Escherichia coli and Saccharomyces cerevisiae transcriptomes, we conducted a genome-scale study aiming at dissecting the determinants of translation efficiency. There is a significant association between codon bias and translation efficiency across all endogenous genes in E. coli and S. cerevisiae but no association between folding energy and translation efficiency, demonstrating the role of codon bias as an important determinant of translation efficiency. However, folding energy does modulate the strength of association between codon bias and translation efficiency, which is maximized at very weak mRNA folding (i.e., high folding energy) levels. We find a strong correlation between the genomic profiles of ribosomal density and genomic profiles of folding energy across mRNA, suggesting that lower folding energies slow down the ribosomes and decrease translation efficiency. Accordingly, we find that selection forces act near uniformly to decrease the folding energy at the beginning of genes. In summary, these findings testify that in endogenous genes, folding energy affects translation efficiency in a global manner that is not related to the expression levels of individual genes, and thus cannot be detected by correlation with their expression levels.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. - PubMed
-
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. - PubMed
-
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. - PubMed
-
- Parmley JL, Hurst LD. How do synonymous mutations affect fitness? BioEssays. 2007;29:515–519. - PubMed
-
- Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

