Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
- PMID: 20133651
- PMCID: PMC2840367
- DOI: 10.1073/pnas.0906101107
Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Abstract
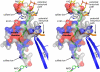
The bifunctional proline catabolic flavoenzyme, proline utilization A (PutA), catalyzes the oxidation of proline to glutamate via the sequential activities of FAD-dependent proline dehydrogenase (PRODH) and NAD(+)-dependent Delta(1)-pyrroline-5-carboxylate dehydrogenase (P5CDH) domains. Although structures for some of the domains of PutA are known, a structure for the full-length protein has not previously been solved. Here we report the 2.1 A resolution crystal structure of PutA from Bradyrhizobium japonicum, along with data from small-angle x-ray scattering, analytical ultracentrifugation, and steady-state and rapid-reaction kinetics. PutA forms a ring-shaped tetramer in solution having a diameter of 150 A. Within each protomer, the PRODH and P5CDH active sites face each other at a distance of 41 A and are connected by a large, irregularly shaped cavity. Kinetics measurements show that glutamate production occurs without a lag phase, suggesting that the intermediate, Delta(1)-pyrroline-5-carboxylate, is preferably transferred to the P5CDH domain rather than released into the bulk medium. The structural and kinetic data imply that the cavity serves both as a microscopic vessel for the hydrolysis of Delta(1)-pyrroline-5-carboxylate to glutamate semialdehyde and a protected conduit for the transport of glutamate semialdehyde to the P5CDH active site.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35(4):681–690. - PubMed
-
- Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and Molecular Basis of Inherited Disease. New York: McGraw Hill; 2001. pp. 1821–1838.
-
- Willis A, Bender HU, Steel G, Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008;35(4):673–679. - PubMed
-
- Nagata K, et al. L-Serine, D- and L-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology+ 2003;149(Pt 8):2023–2030. - PubMed
-
- Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149(1):1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

