Long-range enhancers on 8q24 regulate c-Myc
- PMID: 20133699
- PMCID: PMC2840341
- DOI: 10.1073/pnas.0906067107
Long-range enhancers on 8q24 regulate c-Myc
Abstract
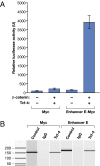
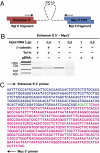
Recent genomewide association studies have found multiple genetic variants on chromosome 8q24 that are significantly associated with an increased susceptibility to prostate, colorectal, and breast cancer. These risk loci are located in a "gene desert," a few hundred kilobases telomeric to the Myc gene. To date, the biological mechanism(s) underlying these associations remain unclear. It has been speculated that these 8q24 genetic variant(s) might affect Myc expression by altering its regulation or amplification status. Here, we show that multiple enhancer elements are present within this region and that they can regulate transcription of Myc. We also demonstrate that one such enhancer element physically interacts with the Myc promoter via transcription factor Tcf-4 binding and acts in an allele specific manner to regulate Myc expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Cher ML, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56:3091–3102. - PubMed
-
- Visakorpi T, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. - PubMed
-
- Korsmeyer SJ. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu Rev Immunol. 1992;10:785–807. - PubMed
-
- Douglas EJ, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. - PubMed
-
- Ribeiro FR, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–3970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

