Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine
- PMID: 20133752
- PMCID: PMC2840323
- DOI: 10.1073/pnas.0914869107
Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine
Abstract
Modification of the cytidine in the first anticodon position of the AUA decoding tRNA(Ile) (tRNA2(Ile)) of bacteria and archaea is essential for this tRNA to read the isoleucine codon AUA and to differentiate between AUA and the methionine codon AUG. To identify the modified cytidine in archaea, we have purified this tRNA species from Haloarcula marismortui, established its codon reading properties, used liquid chromatography-mass spectrometry (LC-MS) to map RNase A and T1 digestion products onto the tRNA, and used LC-MS/MS to sequence the oligonucleotides in RNase A digests. These analyses revealed that the modification of cytidine in the anticodon of tRNA2(Ile) adds 112 mass units to its molecular mass and makes the glycosidic bond unusually labile during mass spectral analyses. Accurate mass LC-MS and LC-MS/MS analysis of total nucleoside digests of the tRNA2(Ile) demonstrated the absence in the modified cytidine of the C2-oxo group and its replacement by agmatine (decarboxy-arginine) through a secondary amine linkage. We propose the name agmatidine, abbreviation C(+), for this modified cytidine. Agmatidine is also present in Methanococcus maripaludis tRNA2(Ile) and in Sulfolobus solfataricus total tRNA, indicating its probable occurrence in the AUA decoding tRNA(Ile) of euryarchaea and crenarchaea. The identification of agmatidine shows that bacteria and archaea have developed very similar strategies for reading the isoleucine codon AUA while discriminating against the methionine codon AUG.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 from H. marismortui. Location of modified nucleosides is based on Gupta (46) and LC-MS analysis presented in this work. G+ (archaeosine);
from H. marismortui. Location of modified nucleosides is based on Gupta (46) and LC-MS analysis presented in this work. G+ (archaeosine);  (N2, N2-dimethylguanosine); C* (modified cytidine); t6A (N6-threonylcarbamoyladenosine); m5C (5-methylcytidine); m1ψ (1-methylpseudouridine); ψ (pseudouridine); Cm (2′-O-methylcytidine); m1I (1-methylinosine). (B) Purification of isoleucine tRNAs from H. marismortui. Native PAGE analysis of total tRNA, purified
(N2, N2-dimethylguanosine); C* (modified cytidine); t6A (N6-threonylcarbamoyladenosine); m5C (5-methylcytidine); m1ψ (1-methylpseudouridine); ψ (pseudouridine); Cm (2′-O-methylcytidine); m1I (1-methylinosine). (B) Purification of isoleucine tRNAs from H. marismortui. Native PAGE analysis of total tRNA, purified  (Ile2) and
(Ile2) and  (Ile1) is shown. tRNAs are visualized by ethidium bromide staining or Northern blot analysis using probes specific for
(Ile1) is shown. tRNAs are visualized by ethidium bromide staining or Northern blot analysis using probes specific for  and
and  as indicated. (C) Characterization of purified
as indicated. (C) Characterization of purified  . The homogeneity of
. The homogeneity of  was confirmed by partial RNase T1 digest (lane T1) of 5′-32P-labeled tRNA. 32P-labeled fragments were separated by denaturing PAGE and visualized by autoradiography; lane OH-, partial alkali digest.
was confirmed by partial RNase T1 digest (lane T1) of 5′-32P-labeled tRNA. 32P-labeled fragments were separated by denaturing PAGE and visualized by autoradiography; lane OH-, partial alkali digest.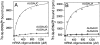
 and
and  . Template-dependent binding of purified
. Template-dependent binding of purified  (A) and
(A) and  (B) to ribosomes isolated from H. marismortui. Oligonucleotides used were AUGAUG (▵), AUGAUA (□) and AUGAUC (○).
(B) to ribosomes isolated from H. marismortui. Oligonucleotides used were AUGAUG (▵), AUGAUA (□) and AUGAUC (○).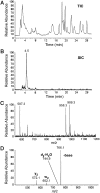
 . (A) TIC of RNase A/BAP digestion of H. marismortui
. (A) TIC of RNase A/BAP digestion of H. marismortui  . (B) SIC of the anticodon-derived trinucleoside diphosphate 5′-C*pApU-3′ at m/z 989.3. (C) Mass spectrum of oligonucleotides eluting at 4.5 min. (D) MS/MS analysis of the trinucleoside diphosphate at m/z 989.3 (22).
. (B) SIC of the anticodon-derived trinucleoside diphosphate 5′-C*pApU-3′ at m/z 989.3. (C) Mass spectrum of oligonucleotides eluting at 4.5 min. (D) MS/MS analysis of the trinucleoside diphosphate at m/z 989.3 (22).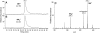
 was digested to nucleosides and analyzed by LC-UV-MS (for complete analysis see
was digested to nucleosides and analyzed by LC-UV-MS (for complete analysis see  ). (C) Mass spectrum of the modified cytidine C*; C* coelutes with the leading edge of archaeosine.
). (C) Mass spectrum of the modified cytidine C*; C* coelutes with the leading edge of archaeosine.
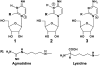
 to lysidine present in the corresponding bacterial tRNA. Possible tautomeric structures of protonated agmatidine and lysidine are shown. Tautomers 1 and 2 could base pair with A.
to lysidine present in the corresponding bacterial tRNA. Possible tautomeric structures of protonated agmatidine and lysidine are shown. Tautomers 1 and 2 could base pair with A.References
-
- Crick FH. Codon-anticodon pairing: The wobble hypothesis. J Mol Biol. 1966;19:548–555. - PubMed
-
- Söll D, RajBhandary UL. Studies on polynucleotides. LXXVI. Specificity of transfer RNA for codon recognition as studied by amino acid incorporation. J Mol Biol. 1967;29:113–124. - PubMed
-
- Harada F, Nishimura S. Purification and characterization of AUA specific isoleucine transfer ribonucleic acid from Escherichia coli B. Biochemistry. 1974;13:300–307. - PubMed
-
- Muramatsu T, et al. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988;263:9261–9267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

